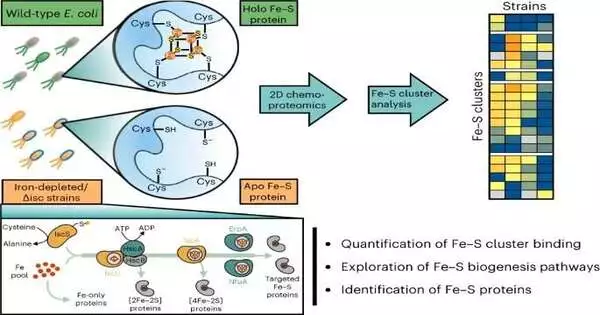
Boston School physicists have fostered a methodology to screen for the presence or absence of iron-sulfur groups, which are vital for the capability of different proteins; the group is detailed as of late in the journal Nature Substance Science.
According to co-creators of the review, Teacher of Science Eranthie Weerapana and Senior Exploration Partner Daniel Bak, the iron-sulfur (Fe-S) proteomic stage the group created allowed them to gain a more significant understanding into the elements of iron-sulfur bunch conveyance and restriction.
Iron-sulfur bunches are found in proteins that are vital for human wellbeing (for example, DNA fix chemicals) and significant in the energy sciences for biofuel cells and cleaner modern cycles.
Regardless, the investigation of iron-sulfur clusters has always been complicated by the unsteadiness and elements of the Fe-S group protein, according to Bak.
“By mining the datasets we generated, we were able to construct a list of potentially novel Fe-S proteins, two of which were validated as iron-sulfur cluster binding proteins.”
Professor of Chemistry Eranthie Weerapana
“We set off to foster an innovation that would permit us to worldwide evaluate the elements of Fe-S bunch restricting in a whole proteome without the requirement for relentless protein cleansing, radioisotopes of iron or sulfur, or past information on bunch restricting destinations,” Bak said.
Numerous proteins contain metal cofactors, for example, iron-sulfur bunches, which he made sense of. These inorganic cofactors consider novel protein capabilities, for example, one-electron redox science, that are not accessible to the 20 standard protein amino acids.
The group played out the concentrate in the microscopic organism E. coli, which is a model life form with a very well-portrayed iron-sulfur proteome. According to Weerapana, the group used advanced mass spectrometry-based proteomic methodologies combined with compound tests that can separate between the bunch-bound and -unbound types of a protein to quantify the limiting of Fe-S bunches to protein focuses.
The researchers discovered a differential awareness of iron-sulfur bunches to both iron limitation and debilitation of the pathways E. coli uses to orchestrate Fe-S groups, recommending a prioritization of iron-sulfur bunch conveyance in E. coli. Information on iron-sulfur group prioritization can provide insight into how various microorganisms can absorb and thrive in natural environments where iron is scarce.
The specialists were likewise ready to separate the jobs of the different iron-sulfur biosynthetic compounds, some of which are fundamental for the conveyance of practically all Fe-S groups, while others assume more unambiguous parts in the conveyance of bunches to a particular subset of Fe-S proteins.
“Mining the datasets that we created, we could gather a rundown of possibly original Fe-S proteins, two of which were approved as iron-sulfur group restricting proteins,” said Weerapana. “This opens the chance of utilizing proteomics to recognize novel Fe-S group science not just in E. coli, but in different life forms too.”
The analysts expressed that the subsequent stage is to make an interpretation of this work for different creatures and model frameworks, including cell culture models of FeS-subordinate human infections like Friedrich’s ataxia. The new stage offers an amazing asset to portray components of Fe-S group combination and conveyance as well as distinguish new iron-sulfur-subordinate cell pathways.
More information: Daniel W. Bak et al, Monitoring Fe–S cluster occupancy across the E. coli proteome using chemoproteomics, Nature Chemical Biology (2023). DOI: 10.1038/s41589-022-01227-9