Alzheimer’s disease (promotion) is a complex neurodegenerative disease with hereditary and ecological starting points. While males have higher mortality rates, females experience faster cognitive decline and cerebral atrophy. Utilizing another AI technique they created called “Transformative Activity AI (EAML),” scientists at Baylor School of Medication and the Jan and Dan Duncan Neurological Exploration Organization (Duncan NRI) at Texas Youngsters’ Emergency Clinic have found sex-explicit qualities and sub-atomic pathways that add to the turn of events and movement of this condition. Nature Communications published the study.
Dr. Olivier Lichtarge, MD, Ph.D., professor of biochemistry and molecular biology at Baylor College of Medicine, stated, “We have developed a unique machine-learning software that uses an advanced computational predictive metric called the evolutionary action (EA) score as a feature to identify genetic factors that influence AD risk separately in males and females.” We can now probe smaller cohorts with greater precision and identify genes involved in sex-specific differences in AD thanks to this strategy, which makes efficient use of a huge amount of evolutionary data.”
Non-synonymous coding variants are defined as DNA mutations that affect the structure and function of the resulting protein. EAML is an ensemble computational approach that uses the evolutionary action (EA) score to estimate the deleterious effect of non-synonymous coding variants on biological processes.
“We created a one-of-a-kind machine-learning software that employs an advanced computational predictive metric known as the evolutionary action (EA) score as a feature to identify genetic factors that influence AD risk separately in males and females,”
Dr. Olivier Lichtarge, MD, Ph.D., professor of biochemistry and molecular biology at Baylor College of Medicine,
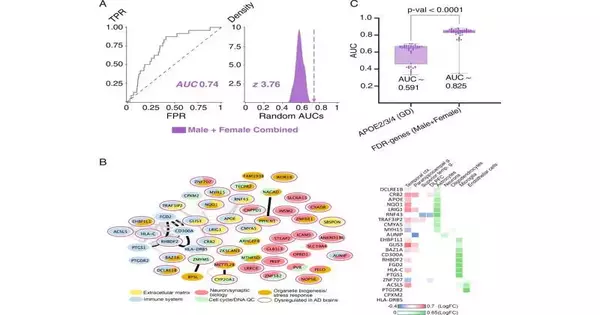
Lichtarge and colleagues identified 98 genes associated with AD by analyzing coding variants in 2,729 AD patients and 2,441 control subjects using EAML. The general value of combining a machine-learning approach with the phylogenetic evolutionary information contained in EA to identify genes and pathways linked to a complex disease like AD was supported by these, which included several genes that are known to play a significant role in AD biology. They also discovered that these genes were expressed abnormally in AD brains and that they made functional connections. Microglial and astrocytic biology, as well as mediated pathways for neuroinflammation, may play a role in AD pathophysiology through specific pathways.
Then, they worked together with Dr. Ismael Al-Ramahi, Dr. Juan Botas, and their groups at the Middle for Alzheimer’s and Neurodegenerative Infections and Duncan NRI to test the homologs of the 98 EAML up-and-comers’ qualities utilizing two natural product fly models of promotion. For this, they utilized a robot-aided, cutting-edge social testing stage, which considers high-throughput separates vivo. They found 36 qualities adjusted tau-initiated degeneration and 29 qualities regulated A42-instigated neurodegeneration. These included nine genes that could slow down the neurodegeneration caused by both Tau and A42, two proteins that are known to build up in people with AD. This highlighted potential therapeutic avenues that could be gained by targeting these genes and strongly validated the functional involvement of the identified candidates in mediating in vivo neurodegeneration.
They then applied EAML analysis separately to male and female participants in this cohort because the objective of this study was to comprehend how AD manifests and progresses differently in men and women. They found 157 promotion-related qualities in guys and 127 in females. The qualities distinguished in this sex-isolated study were viewed as more firmly associated with known promotion GWAS qualities than those recognized in the consolidated sex studies. These discoveries recommend that sex-isolated investigation expands the responsiveness of distinguishing promotion-related qualities and further develops risk-apprehension capacity.
In addition, they discovered that one sex’s AD development may be influenced more strongly by particular biological pathways. For instance, a module related to cell cycle control and DNA quality control was found to be involved in female-specific EAML candidates. We were thrilled to discover a group of genes that were associated with BRCA1, a gene that has been linked to breast cancer, and were neuroprotective in females. These discoveries recommend possible organic associations among promotion and bosom malignant growth, two sicknesses that are more regular in females than guys.” Added Dr. Ismael Al-Ramahi. The design of AD-specific sex-stratified clinical trials and the development of therapeutic strategies could greatly benefit from these findings.
Also, even when the team tested it with smaller sample sizes, EAML kept its ability to predict with solid, consistent targets. Even with only 700 samples, EAML was able to recover more than half of the candidates in the entire data set, which is a significant improvement over the predictive algorithms that are currently in use. The authors believe that researchers will be able to use smaller data sets to make accurate and reliable predictions with this significantly improved capability. This will open the door to incorporating sex-specific analyses into disease-gene association studies that may not have produced reliable results using established methods.
Dr. Juan Botas, a professor in the Baylor Department of Molecular and Human Genetics, added, “Our success in using EAML to find new targets for AD not only provides a fresh perspective on the genetic factors influencing this disorder but also underscores the importance of systematically applying sex-specific analyses when studying disease-gene associations.” This imaginative methodology can possibly change how we might interpret complex sicknesses like promotion and drive the advancement of customized therapies custom-made to every individual’s hereditary cosmetics.”
Thomas Bourquard, Kwanghyuk Lee, Minh Pham, Dillon Shapiro, Yashwanth Lagisetty, Sharin Soleimani, Samantha Mota, Kevin Wilhelm, Maryam Samieinasab, Young Won Kim, Eunna Huh, Jennifer Asmussen, and Panagiotis Katsonis are some of the other people who were involved in the study. They are a part of one of the following organizations: Jan and Dan Duncan Neurological Research Institute at Texas Children’s Hospital, Baylor College of Medicine, and UTHealth McGovern Medical School.
More information: Thomas Bourquard et al, Functional variants identify sex-specific genes and pathways in Alzheimer’s Disease, Nature Communications (2023). DOI: 10.1038/s41467-023-38374-z