Super-resolution microscopy techniques are fundamental for revealing the designs of cells and the elements of atoms. Since specialists conquered the goal’s furthest reaches of around 250 nanometers (while winning the 2014 Nobel Prize in Science for their endeavors), which had for quite some time been viewed as outright, the techniques for microscopy have advanced quickly.
Currently, a group led by LMU scientist Prof. Philip Tinnefeld has made further development through the mix of different techniques, accomplishing the most elevated goal in three-layered space and making ready for a generally new methodology for quicker imaging of thick sub-atomic designs. The new strategy sets a pivotal goal of under 0.3 nanometers.
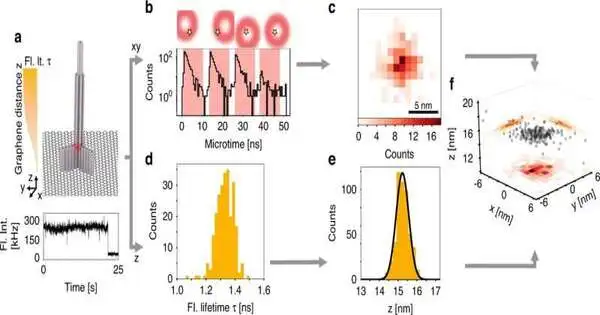
The scientists joined the alleged pMINFLUX strategy created by Tinnefeld’s group with a methodology that uses the extraordinary properties of graphene as an energy acceptor. pMINFLUX depends on the estimation of the fluorescence force of particles energized by laser beats. The technique makes it conceivable to recognize their sidelong distances with a goal of only 1 nanometer.
“The combination of GET-pMINFLUX and L-PAINT allows us to examine molecular structures and dynamics that are crucial to our knowledge of biomolecular events in cells.”
LMU chemist Prof. Philip Tinnefeld
Graphene retains the energy of a fluorescent particle that is something like 40 nanometers away from its surface. The fluorescence power of the particle consequently depends upon its separation from graphene and can be utilized for pivotal distance estimation.
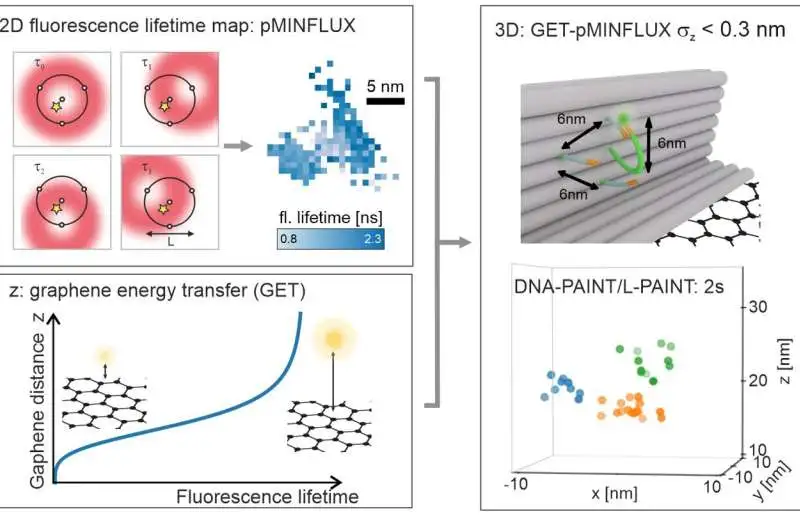
a, pMINFLUX investigates the place of a fluorophore with numerous spatially uprooted donut pillars and yields 2D fluorescence lifetime pictures with nanometer accuracy. b, Graphene gives an action to the pivotal distance of graphene. The fluorescence lifetime abbreviates the closer a fluorophore is to graphene. c. Joining the parallel data of pMINFLUX with the hub graphene distance data yields 3D limitations. GET-pMINFLUX yields photon-effective limitations with nanometer accuracy. This empowers L-PAINT. The schematic of the DNA origami structure has a DNA pointer projecting. The fluorophore-altered DNA-pointer can fleetingly travel to one of three restricting locales separated by 6 nm. Inside 2, this thick construction is constrained to nanometer accuracy in 3D by consolidating L-PAINT and GET-pMINFLUX. Credit: by Jonas Zähringer, Fiona Cole, and Johann Bohlen Florian Steiner, Izabela Kamiska, and Philip Tinnefeld
DNA-PAINT speeds up
Thusly, the mix of pMINFLUX with this supposed graphene energy move (GET) outfits data about sub-atomic distances in each of the three aspects — and does this at the most elevated goal reachable to date of under 0.3 nanometers. “The high accuracy of GET-pMINFLUX makes the way for new methodologies for further developing super-goal microscopy,” says Jonas Zähringer, lead author of the paper.
The specialists additionally utilized this to speed up super-resolution microscopy. To this end, they focused on DNA nanotechnology to foster the supposed L-PAINT approach. As opposed to DNA-PAINT, a procedure that empowers super-goals through the limiting and unbinding of a DNA strand named with a fluorescent color, the DNA strand in L-PAINT has two restricting successions.
Furthermore, the scientists planned a limiting ordered progression to such an extent that the L-PAINT DNA strand ties longer on one side. This permits the opposite finish of the strand to locally filter the particle positions at a fast rate.
“As well as speeding up, this allows the checking of thick groups quicker than the bends emerging from warm float,” says Tinnefeld. “Our blend of GET-pMINFLUX and L-PAINT empowers us to examine designs and elements at the sub-atomic level that are essential to how we might interpret biomolecular responses in cells.”
The discoveries are distributed in the book Light: Science and Applications.
More information: Jonas Zähringer et al, Combining pMINFLUX, graphene energy transfer and DNA-PAINT for nanometer precise 3D super-resolution microscopy, Light: Science & Applications (2023). DOI: 10.1038/s41377-023-01111-8