With a major helping hand from man-made reasoning and a weighty portion of human touch, Tim Cernak’s lab at the College of Michigan has made a disclosure that decisively accelerates the tedious compound course of building particles that are destined to be the upcoming drugs, agrichemicals, or materials.
The revelation, distributed in the Feb. 3 issue of Science, is the climax of long periods of compound blend and information science research by the Cernak Lab in the School of Drug Store and Branch of Science.
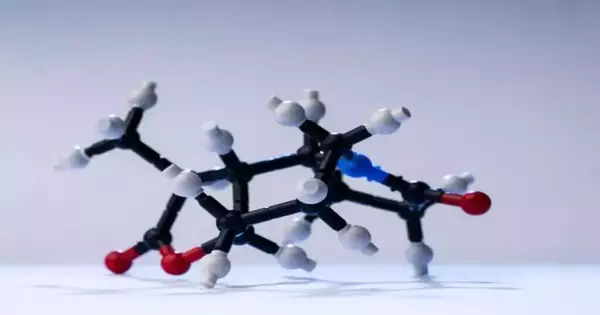
The objective of the examination was to recognize key responses in the union of a particle, thereby lessening the cycle to as few steps as could be expected. Eventually, Cernak and his group accomplished the blend of a perplexing alkaloid tracked down in nature in only three stages. Past blends had been somewhere in the range of seven and 26 strides.
“Making a chemical structure with atoms in just the correct location to give you effective and harmless drugs, for example, is difficult. It necessitates a chemical synthesis technique based on chemical building blocks that may be purchased and then stitched together utilizing chemical reactions.”
Cernak, assistant professor of medicinal chemistry and chemistry.
“Making a compound design that has iotas in the perfect spot to give you solid and nontoxic medications, for example, is precarious,” said Cernak, a partner teacher of restorative science and science. “It necessitates a compound blend system based on synthetic structure blocks that can be purchased and then joined together utilizing substance responses.”
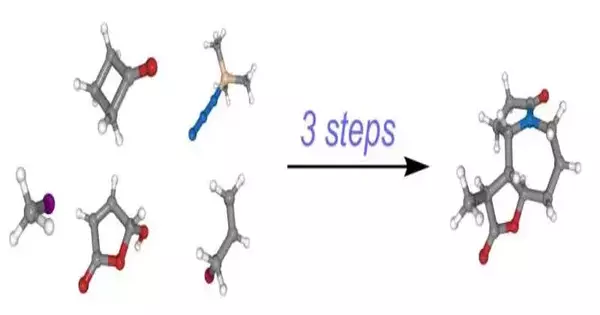
Tim Cernak and his group copied a perplexing particle in just three stages.
The achievement has strong ramifications for accelerating the improvement of medications.
Cernak compared the development of these intricate atoms to playing chess. You want to organize a progression of moves to get to the furthest limit of the game. While there’s a near-endless number of potential moves, there’s a rationale that can be followed.
“We fostered a rationale here, based on chart hypotheses, to get as far as possible as quickly as possible,” he explained.
Cernak and his partners utilized Synthesis Retrosynthesis Programming, which furnishes researchers with a data set of pathways, or steps, and recipes for a great many sub-atomic designs. This gave the group a huge amount of computational union information to play with.
Di Wang (from left), Rui Zhang, Tim Cernak, and Yingfu Lin in the Cernak Lab at the Science Building.
Utilizing a calculation they created to arrange the information, the scientists recognized the means along the pathway that were high effect, or key stages, and the means that were gaining ground toward finishing the blend in any case wasteful for the entire cycle.
“We trust this exploration can prompt better medicines,” Cernak said. “Up to this point, we have been restricted in the atomic designs we can rapidly access with compound union.”
Co-creators include Yingfu Lin, a senior research individual in pharmaceuticals; Rui (Sam) Zhang, a doctoral understudy in science; and Di Wang, a doctoral understudy in pharmaceuticals.
More information: Yingfu Lin et al, Computer-aided key step generation in alkaloid total synthesis, Science (2023). DOI: 10.1126/science.ade8459