Naturally cluttered proteins (IDPs) are generally found in the proteomes of eukaryotes and assume key roles in life cycles, like records of hereditary data and flagging. Aside from being generally exceptionally dull, hydrophilic, and electrically charged, as well as encoding straightforward groupings of qualities, IDPs additionally separate themselves in their normal overflow and primary viewpoints, which become the premise of the “jumble capability worldview” of proteins.
Throughout the course of many years, the job of IDPs in human illnesses and as medication targets has been effectively considered, while how to describe the exceptionally adaptable and heterogeneous conformities of IDPs at high goals has remained a central question in this field.
Over 15% of IDP atoms are layer-bound in cells, and their inside elements and general (translational and rotational) movements inside the phospholipid bilayers are firmly connected with their physicochemical properties and organic capabilities; however, these unique cycles are hard to catch and quantitatively described by ordinary underlying examination techniques.
A gathering of specialists led by Prof. Long Dong from the College of Science and Innovation of China (USTC) has fostered an IDP spectroscopy strategy in view of the layer paramagnetic unwinding upgrade (mPRE) procedure, which has effectively accomplished high-accuracy displaying of the inside compliances, directions, and submersion profundities of IDPs. The outcomes have been distributed in the Diary of the American Synthetic Culture.
In this work, the scientists investigated exhaustively the adaptability and portability of the twist test particles inside the film for exact translation of the mPRE ghastly information and proposed a weighted three-layered (3D) lattice model in light of all-iota reproductions for quantitatively depicting the impact of the twist test elements on the layer paramagnetic unwinding upgrade rate.
Exploiting the high computational effectiveness of the model, the specialists further fostered a calculation that is definite in the supporting data (SI) for advancing directions of the worldwide and inward levels of opportunity of movement of layer-bound IDPs by means of the superposition of z-organizes just, custom fitted for the mPRE information examination, and developed an all-particle group model of IDP in a verifiable film climate.
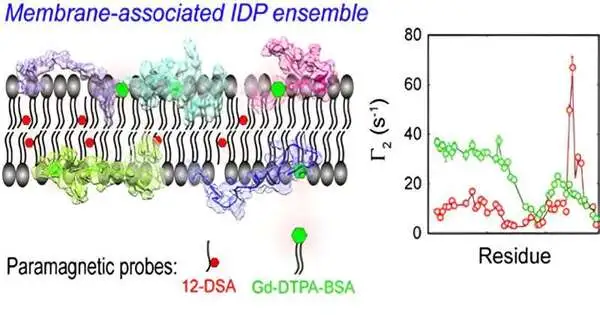
CD3ε is a part of the lymphocyte receptor (TCR) complex responsible for white blood cell antigen acknowledgment. The CD3ε cytoplasmic space (CD3εCD) contains immunoreceptor tyrosine-based enactment themes (ITAMs), and it frames a fluffy complex with lipid bilayers in a naturally scattered state and controls the flagging action of the fluffy complex by using dynamic film safeguarding of key tyrosine destinations.
The scientists settled the troupe in light of sub-atomic elements of CD3εCD in lipid bilayers by applying an all-particle outfit model arrangement of IDP in a certain layer climate.
The gathering from mPRE exploratory boundaries maps the unique circulation of CD3εCD in various locales of the film at the nuclear level and uncovers key contrasts in the layer cooperations of various tyrosine destinations on ITAM, giving another unthinking clarification to the monophosphorylation example of ITAM.
The mPRE spectroscopic examination technique laid out in this work is supposed to extensively work with nuclear goal investigations of different practical film IDPs.
More information: Hong Jin et al. Quantitative Ensemble Interpretation of Membrane Paramagnetic Relaxation Enhancement (mPRE) for Studying Membrane-Associated Intrinsically Disordered Proteins, Journal of the American Chemical Society (2023). DOI: 10.1021/jacs.3c10847