Ongoing neuroscience studies reliably illustrate the role of the C9ORF72 quality in the advancement of a few neurodegenerative illnesses. These investigations discovered that changes of this quality increase the risk of creating amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), two neurodegenerative problems described by engine impedances, issues with conveying, and other particular side effects.
Scientists at Johns Hopkins College Institute of Medication, the College of Chicago, the Howard Hughes Clinical Foundation, and different organizations in the US have recently completed a review focused on better understanding the cycles through which C9ORF72 quality transformation could at last contribute to the improvement of ALS and FTD. Their discoveries, published in Nature Neuroscience, feature a promising road that could work on restorative mediations for these complex neurodegenerative problems.
“My examination bunch has been keen on recognizing modifiers of the C9ORF72 rehash RNA digestion,” Dr. Shuying Sun, one of the analysts who did the review, told Clinical Xpress. “A hexanucleotide rehash of development in the C9ORF72 quality is the most pervasive hereditary reason for the two ALS and FTD. As the extension is situated in the noncoding locale, the rehash containing RNA is accepted to be one central point that drives the sickness pathogenesis.”
“The most common genetic cause of ALS and FTD is a hexanucleotide repeat expansion in the C9ORF72 gene. Because the growth occurs in the noncoding area, repeat-containing RNA is thought to be a substantial component in illness etiology.”
Dr. Shuying Sun, one of the researchers who carried out the study, told Medical Xpress.
Sun and her partners have been attempting to recognize hereditary modifiers of the unusual creation of poisonous poly-dipeptide proteins from the recurrent RNA. This could thus permit them to recognize feasible techniques to forestall the gathering of these harmful proteins and hence lessen their poisonous impacts. In one of their past examinations, they played out an extensive CRISPR-Cas9 screen to look for hereditary modifiers of dipeptide creation.
“We found that the two m6A methyltransferases have up-and-coming qualities that can regulate the poly-dipeptide level,” Sun made sense of. “We likewise dissected public proteomic information and found the two methyltransferases are downregulated in C9ORF72-ALS/FTD patient iPS-neurons. By and large, this started our further concentration on the m6A dysregulation and how this adds to neurodegeneration in C9ORF72-ALS/FTD.”
Sun and her partners played out a progression of investigations on tolerant inferred incited pluripotent undeveloped cell (iPSC)-separated neurons (iPSNs) and posthumous tissues removed from expired patients with ALS/FTD and with C9ORF72 quality transformations. Utilizing different high-throughput hereditary sequencing strategies, they explicitly evaluated an RNA alteration known as M6A (methyladenosine) and its effect on the guideline of qualities across the full scope of communicated messenger RNA (mRNA), known as the transcriptome.
“We likewise inspected the atomic component of the m6A guideline on C9 rehash RNA,” Sun said. “At long last, we estimated neuron endurance to inspect the salvage viability by adjusting the m6A pathway. Our discoveries uncover an original layer of RNA guidelines that assume a basic part in the pathogenic system: fundamental neurodegeneration.”
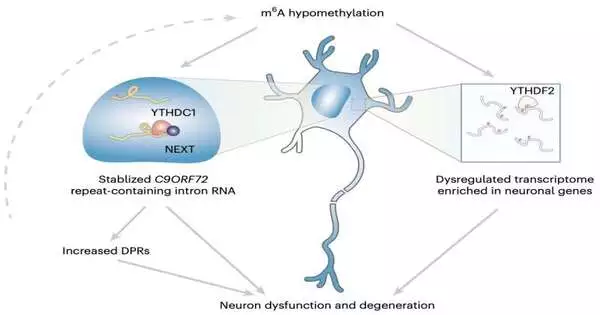
Generally, the discoveries assembled by this exploration group showed that the m6A adjustment in the C9ORF72 hereditary succession of extended rehashes worked with the rot of RNA by means of the atomic peruser YTHDC1, hence directing the rehashes. Also, the decrease in m6A seemed to work with the amassing of rehash RNAs and the encoding of harmful proteins known to be related to neurodegeneration.
By raising m6A methylation, Sun and her partners had the option to fundamentally diminish the degree of rehash RNAs, defending the neurons they were investigating and lessening their risk of rotting. Later on, these outcomes could make way for the improvement of additional powerful medicines for ALS and FTD, for example, by inciting preliminary surveys to assess the worth of epitranscriptomic treatment as a likely intervention.
“We might want to further test the salvage viability in vivo utilizing fitting mouse models,” Sun added. “We are additionally keen on creating drug approaches focusing on this pathway. Moreover, we desire to explore the fundamental illness components, including RNA digestion and m6A dysregulation, in irregular ALS and other neurological problems.”
More information: Yini Li et al, Globally reduced N6-methyladenosine (m6A) in C9ORF72-ALS/FTD dysregulates RNA metabolism and contributes to neurodegeneration, Nature Neuroscience (2023). DOI: 10.1038/s41593-023-01374-9