A new study has discovered that genomic loops do not endure long in cells, suggesting that theories about how loops govern gene expression may need to be altered.
Proteins cover DNA in human chromosomes to form an extremely long beaded thread. This “string” is folded into multiple loops, which are thought to assist cells in controlling gene expression and aid DNA repair, among other things. According to a new MIT study, these loops are much more dynamic and shorter-lived than previously imagined.
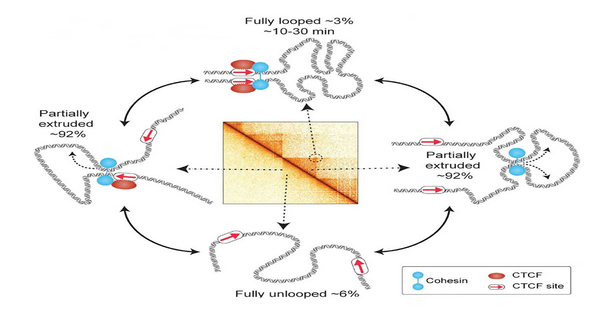
In the latest study, researchers were able to track the movement of one stretch of the genome in a living cell for roughly two hours. They discovered that this segment was only fully looped 3 to 6% of the time, with the loop lasting only 10 to 30 minutes. According to the researchers, the findings suggest that scientists’ present understanding of how loops regulate gene expression may need to be changed.
Many models in the field have been images of static loops that regulate these activities. Our recent work demonstrates that this picture is not entirely accurate. MIT’s Underwood-Prescott Career Development Assistant Professor of Biological Engineering, Anders Sejr Hansen, agrees. “We believe these areas’ functional states are substantially more dynamic.”
Hansen is one of the study’s senior authors, along with Leonid Mirny, an MIT Institute for Medical Engineering and Science and Department of Physics professor, and Christoph Zechner, a group leader at the Max Planck Institute of Molecular Cell Biology and Genetics in Dresden, Germany, and the Center for Systems Biology Dresden. The paper’s principal authors are MIT postdoc Michele Gabriele, recent Harvard University Ph.D. recipient Hugo Brando, and MIT graduate student Simon Grosse-Holz.
Out of the loop.
Using computer simulations and experimental data, scientists, including Mirny’s lab at MIT, demonstrated that loops in the genome are produced via an extrusion process in which a molecular motor encourages the formation of progressively larger loops. When the engine comes across a “stop sign” in DNA, it comes to a halt. The motor that extrudes such loops is a protein complex called cohesin, and the stop sign is the DNA-bound protein CTCF. Previously, cohesin-mediated loops between CTCF sites were observed.
Those trials, however, only provided a snapshot of a single point in time, with no indication of how the loops alter over time. In their latest study, the researchers created strategies for fluorescently labeling CTCF DNA locations, allowing them to observe the DNA loops over several hours. They also developed a new computational strategy for inferring looping events from imaging data.
“This strategy was critical for distinguishing signal from noise in our experimental data and quantifying looping,” Zechner explains. “We feel that as we continue to push the limits of detection with tests, such approaches will become increasingly relevant for biology.”
“Many models in the field have been these pictures of static loops regulating these processes. What our new paper shows is that this picture is not really correct. We suggest that the functional state of these domains is much more dynamic.”
Anders Sejr Hansen, the Underwood-Prescott Career Development Assistant Professor of Biological Engineering at MIT.
The technology was utilized by the researchers to image a region of the genome in mouse embryonic stem cells. “If we place our findings in the perspective of one cell division cycle, which lasts around 12 hours,” Grosse-Holz adds, “the completely formed loop only lives for about 20 to 45 minutes, or about 3 to 6 percent of the time.”
Hansen goes on to say that “we shouldn’t think of this completely looped state as the key regulator of gene expression.””We believe that new models are required to explain how the genome’s 3D shape influences gene expression, DNA repair, and other functional downstream processes.”
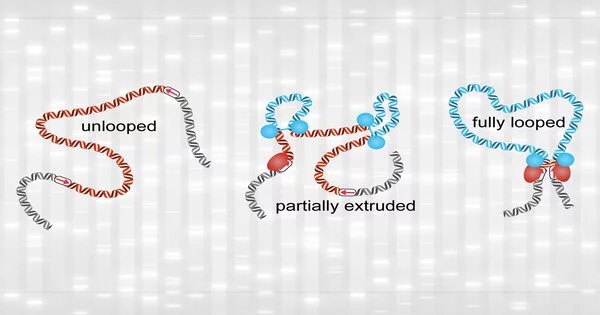
While fully formed loops were uncommon, the researchers discovered that partially extruded loops were present approximately 92% of the time. These tiny loops were difficult to discover using prior approaches for detecting loops in the genome.
In this work, “We were able to measure the relative extents of the unlooped, partially extruded, and fully looped states in this work by merging our experimental data with polymer simulations,” Brando explains.
Because these interactions are brief but frequent, earlier approaches were unable to completely capture their dynamics, Gabriele says. “With our new technique, we can begin to resolve transitions between fully looped and unlooped states.”
The researchers believe that incomplete loops are more significant in gene regulation than completely formed loops. Strands of DNA run along each other as loops form and then disassemble, and these interactions may aid in the discovery of regulatory elements such as enhancers and gene promoters.
“More than 90% of the time, there are some transient loops, and presumably what’s crucial is having those loops that are constantly extruded,” Mirny explains. “The process of extrusion itself may be more essential than the fully looped state, which occurs just briefly.”
More loops to investigate
Because the majority of the other loops in the genome are weaker than the one analyzed in this work, the researchers predict that many additional loops will also prove to be highly transient. They intend to utilize their new technique to investigate some of the other loops in a range of cell types.
“We looked at one of perhaps 10,000 of these loops,” Hansen explains. We have a lot of indirect evidence that the findings are generalizable, but we haven’t proven it. We may begin to tackle other loops in the genome using the technology platform we’ve established, which combines novel experimental and computational methodologies.
The scientists also intend to look at the role of certain circuits in sickness. Many disorders, including FOXG1 syndrome, a neurodevelopmental disability, could be connected to incorrect loop dynamics. The researchers are now investigating how genome loop development affects both the normal and mutant forms of the FOXG1 gene, as well as the cancer-causing gene MYC.
It lasted a long time in cells, suggesting that theories about how loops govern gene expression may need to be altered.
Proteins cover DNA in human chromosomes to form an extremely long beaded thread. This “string” is folded into multiple loops, which are thought to assist cells in controlling gene expression and aid DNA repair, among other things. According to a new MIT study, these loops are much more dynamic and shorter-lived than previously imagined.
In the latest study, researchers were able to track the movement of one stretch of the genome in a living cell for roughly two hours. They discovered that this segment was only fully looped 3 to 6% of the time, with the loop lasting only 10 to 30 minutes. According to the researchers, the findings suggest that scientists’ present understanding of how loops regulate gene expression may need to be changed.
Many models in the field have been images of static loops that regulate these activities. Our recent work demonstrates that this picture is not entirely accurate. MIT’s Underwood-Prescott Career Development Assistant Professor of Biological Engineering, Anders Sejr Hansen, agrees. “We believe these areas’ functional states are substantially more dynamic.”
Hansen is one of the study’s senior authors, along with Leonid Mirny, an MIT Institute for Medical Engineering and Science and Department of Physics professor, and Christoph Zechner, a group leader at the Max Planck Institute of Molecular Cell Biology and Genetics in Dresden, Germany, and the Center for Systems Biology Dresden. The paper’s principal authors are MIT postdoc Michele Gabriele, recent Harvard University Ph.D. recipient Hugo Brando, and MIT graduate student Simon Grosse-Holz.
Out of the loop.
Using computer simulations and experimental data, scientists, including Mirny’s lab at MIT, demonstrated that loops in the genome are produced via an extrusion process in which a molecular motor encourages the formation of progressively larger loops. When the engine comes across a “stop sign” in DNA, it comes to a halt. The motor that extrudes such loops is a protein complex called cohesin, and the stop sign is the DNA-bound protein CTCF. Previously, cohesin-mediated loops between CTCF sites were observed.
Those trials, however, only provided a snapshot of a single point in time, with no indication of how the loops alter over time. In their latest study, the researchers created strategies for fluorescently labeling CTCF DNA locations, allowing them to observe the DNA loops over several hours. They also developed a new computational strategy for inferring looping events from imaging data.
“This strategy was critical for distinguishing signal from noise in our experimental data and quantifying looping,” Zechner explains. “We feel that as we continue to push the limits of detection with tests, such approaches will become increasingly relevant for biology.”
The technology was utilized by the researchers to image a region of the genome in mouse embryonic stem cells. “If we place our findings in the perspective of one cell division cycle, which lasts around 12 hours,” Grosse-Holz adds, “the completely formed loop only lives for about 20 to 45 minutes, or about 3 to 6 percent of the time.”
Hansen goes on to say that “we shouldn’t think of this completely looped state as the key regulator of gene expression.””We believe that new models are required to explain how the genome’s 3D shape influences gene expression, DNA repair, and other functional downstream processes.”
While fully formed loops were uncommon, the researchers discovered that partially extruded loops were present approximately 92% of the time. These tiny loops were difficult to discover using prior approaches for detecting loops in the genome.
In this work, “We were able to measure the relative extents of the unlooped, partially extruded, and fully looped states in this work by merging our experimental data with polymer simulations,” Brando explains.
Because these interactions are brief but frequent, earlier approaches were unable to completely capture their dynamics, Gabriele says. “With our new technique, we can begin to resolve transitions between fully looped and unlooped states.”
The researchers believe that incomplete loops are more significant in gene regulation than completely formed loops. Strands of DNA run along each other as loops form and then disassemble, and these interactions may aid in the discovery of regulatory elements such as enhancers and gene promoters.
“More than 90% of the time, there are some transient loops, and presumably what’s crucial is having those loops that are constantly extruded,” Mirny explains. “The process of extrusion itself may be more essential than the fully looped state, which occurs just briefly.”
More loops to investigate
Because the majority of the other loops in the genome are weaker than the one analyzed in this work, the researchers predict that many additional loops will also prove to be highly transient. They intend to utilize their new technique to investigate some of the other loops in a range of cell types.
“We looked at one of perhaps 10,000 of these loops,” Hansen explains. We have a lot of indirect evidence that the findings are generalizable, but we haven’t proven it. We may begin to tackle other loops in the genome using the technology platform we’ve established, which combines novel experimental and computational methodologies.
The scientists also intend to look at the role of certain circuits in sickness. Many disorders, including FOXG1 syndrome, a neurodevelopmental disability, could be connected to incorrect loop dynamics. The researchers are now investigating how genome loop development affects both the normal and mutant forms of the FOXG1 gene, as well as the cancer-causing gene MYC.