Another review published in Nature reports that an innovation known as spatial omics can be utilized to simultaneously plan the way in which qualities are turned here and there and how they are communicated in various areas of tissues and organs. This superior innovation, created by scientists at Yale College and Karolinska Institutet, could reveal insight into the improvement of tissues as well as on specific infections and how to treat them.
Practically all phones in the body have a similar arrangement of qualities and can, on a basic level, become any sort of cell. What recognizes the cells is the means by which the qualities in our DNA are utilized. Lately, spatial omics have provided us with a more profound understanding of how cells read the genome in exact areas of tissues. Currently, scientists have additionally advanced this innovation to expand information on how tissues create and how various illnesses emerge.
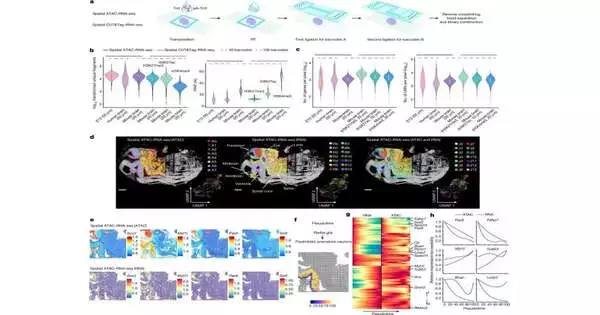
A vital piece of the review is the scientists’ capacity to simultaneously spatially plan two critical parts of our hereditary makeup, the epigenome and the transcriptome. The epigenome controls the systems that turn qualities on and off in individual cells, while the transcriptome is the aftereffect of those quality articulations and what makes every cell one of a kind.
“This has led to some surprising results, providing us with additional insights into how these mechanisms are regulated in different tissue locations and contribute to distinct cell fates.”
Gonçalo Castelo-Branco, professor at the Department of Medical Biochemistry and Biophysics, Karolinska Institutet,
Can distinguish information and result all the while
The epigenome can be considered a breaker box. You can flip the switches, but in the event that you can’t see whether the lights are turning on, your data is restricted. By spatially planning both the epigenome and transcriptome, the specialists fostered an innovation in which both the information (turning on or off a quality) and the result (quality articulation) can be identified in a similar tissue segment. This new innovation gets uncommon bits of knowledge into quality guideline-exact areas in a tissue.
For this review, the analysts adjusted and consolidated their recently evolved methods to plan the epigenome and the transcriptome and applied these strategies to mouse minds and human cerebrum tissue.
“Now that we can join the two, we can see both the systems of how the qualities are turned here and there, as well as the outcome,” says Gonçalo Castelo-Branco, teacher at the Division of Clinical Organic Chemistry and Biophysics at the Karolinska Institutet and one of the comparing creators. “This has driven us to a few startling perceptions, which give us further bits of knowledge into how these cycles are controlled in various tissue regions and add to various cell destinies.”
Propelling the field of customized medication
The work could bring analysts closer to figuring out possible hereditary focuses for drug treatment and propel the field of customized medication.
“In the future, with this innovation, we will actually want to truly comprehend in each and every patient how those disease-advancing and growth-silencer qualities are being directed by the epigenetic components,” says Rong Fan, teacher at Yale College and last creator of the paper. “The entire epigenetic therapeutics field is simply emerging; however, I figure our innovation might possibly engage epigenetic drug disclosure.”
More information: Di Zhang et al, Spatial epigenome–transcriptome co-profiling of mammalian tissues, Nature (2023). DOI: 10.1038/s41586-023-05795-1