Another strategy can enlighten the characters and exercises of cells all through an organ or a growth toward a phenomenal goal, as indicated by a review conducted by specialists at Weill Cornell Medicine, New York-Presbyterian, and the New York Genome Center.
The strategy, portrayed Jan. 2 in a paper in Nature Biotechnology, records quality action designs and the presence of key proteins in cells across tissue tests while holding data about the cells’ exact areas. This empowers the creation of complex, information-rich “maps” of organs, including unhealthy organs and growths, which could be generally valuable in fundamental and clinical exploration.
“This innovation is exciting because it allows us to plan the spatial association of tissues, including cell types, cell exercises, and cell-to-cell communications, as never before,” said study co-senior creator Dr. Dan Landau, an academic partner of medication in the Division of Hematology and Clinical Oncology and a member of the Sandra and Edward Meyer Malignant Growth Community at Weill Cornell Medication and a center employee at the New York Genomics Institute.
“This technology is fascinating because it enables us to map the spatial architecture of tissues, including cell kinds, cell activities, and cell-to-cell connections, in unprecedented detail,”
Dr. Dan Landau, an associate professor of medicine in the Division of Hematology and Medical Oncology.
The other co-senior creator was Dr. Marlon Stoeckius of 10x Genomics, a California-based biotechnology organization that makes lab hardware for the profiling of cells inside tissue tests. Dr. Nir Ben-Chetrit, Xiang Niu, and Ariel Swett, who were each a postdoctoral specialist, graduate understudy, and examination professional in the Landau lab during the review, were the three co-first creators.
The new technique is important for a wide effort by researchers and designers to foster better approaches to “seeing” at small sizes how organs and tissues work. Scientists as of late have made huge advances, especially in methods for profiling quality action and different layers of data in individual cells or small gatherings of cells. Notwithstanding, these procedures regularly require the disintegration of tissues and the partition of cells from their neighbors, so data about profiled cells’ unique areas inside the tissues is lost. The new technique catches that spatial data also, and at a high level.
The Spatial Protein and Transcriptome Sequencing (SPOTS) strategy is based in part on existing 10x Genomics innovation.It utilizes glass slides that are appropriate for imaging tissue tests with conventional magnifying lens-based pathology techniques, but at the same time are covered with a huge number of unique test particles. Every one of the test particles contains a sub-atomic “scanner tag,” signifying its two-layered position on the slide. At the point when a meagerly cut tissue sample is put on the slide and its cells are made penetrable, the test particles on the slide get nearby cells’ courier RNAs (mRNAs), which are basically the records of dynamic qualities. The strategy includes the use of planner antibodies, which target proteins of interest in the tissue and also bind to the exceptional test particles.With quick, computerized strategies, analysts can recognize the caught mRNAs and selected proteins and guide them definitively to their unique areas across the tissue test. The subsequent guides can be viewed alone or contrasted with standard pathology imaging of the example.
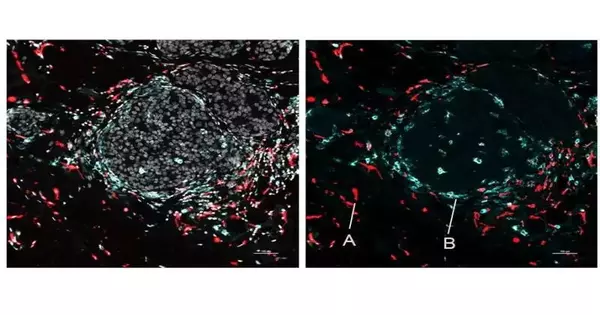
The group exhibited spots on tissue from an ordinary mouse spleen, uncovering the complex useful design of this organ, including bunches of various cell types, their practical states, and how those states shifted with the phones’ areas.
SPOTS was used to plan the phone association of a mouse with breast cancer, demonstrating its potential in malignant growth research.The following guide depicted safe cells called macrophages in two distinct states as indicated by protein markers—one dynamic and growth-fighting, the other resistant suppressive and framing a barrier to cancer protection.
“We could see that these two macrophage subsets are tracked down in various regions of the cancer and communicate with various cells—aand that distinction in microenvironment is logically driving their unmistakable action states,” said Dr. Landau, who is additionally an oncologist at NewYork-Presbyterian/Weill Cornell Clinical Center.
“Such subtleties of the growth invulnerable climate — subtleties that frequently can’t be resolved because invulnerable cells’ meager condition inside cancers — could help with comprehending why some patients respond to resistant treatment and others don’t, and consequently could educate the plan regarding future immunotherapies,” he added.
“This underlying variant of SPOTS has a spatial goal to such an extent that every “pixel” of the subsequent dataset aggregates quality movement data for basically a few cells.” “Be that as it may, the analysts trust they will soon limit this goal to single cells while adding different layers of key cell data,” Dr. Landau said.
More information: Marlon Stoeckius, Integration of whole transcriptome spatial profiling with protein markers, Nature Biotechnology (2023). DOI: 10.1038/s41587-022-01536-3. www.nature.com/articles/s41587-022-01536-3
Journal information: Nature Biotechnology