Proteins comprise the chief structural blocks in every living creature. They are oftentimes depicted as the laborers of the cell, where they—together or individually—play out various fundamental errands. In the case of something turning out badly, the results are frequently serious. Both exploration and medical care have communicated the requirement for viable devices to examine the capabilities and exercises of proteins, and in another article in the logical journal Nature Communications, Professor Ola Söderberg’s group presents MolBoolean, an innovation that is supposed to open significant entryways in cell and disease research.
“MolBoolean” is a strategy to decide the levels of a couple of proteins in individual cells while equally recognizing the levels of these proteins that tight spot one another. “We have kept on fostering a device that we made in 2006, which today is utilized everywhere, and the capabilities that we are currently adding create data about the general measures of various proteins as well as relative measures of protein edifices, hence making the phones’ action status and correspondence more noticeable,” says Ola Söderberg, Professor of Pharmaceutical Cell Biology at Uppsala University. “This can measure up to surveying a café; 10 positive audits give some data, yet it is vital to know whether they depend on ten or 1,000 feelings.”
“Our study intends to develop tools for seeing activities within cells and, for example, to improve understanding of what happens in cancer cells. Knowing the balance of free and interacting proteins is critical for researching cell signaling, and we anticipate MolBoolean will be useful in many research labs.”
Doroteya Raykova
The gathering’s outcomes are arousing extraordinary curiosity in both the scholarly world and industry, and presently the Stockholm-based organization Atlas Antibodies — which as of late surpassed the patent for MolBoolean — is setting up the strategy for the market. The device is supposed to be of extraordinary worth in research and, in the long haul, likewise in medical care to further develop the finding and decision of therapy for disease.
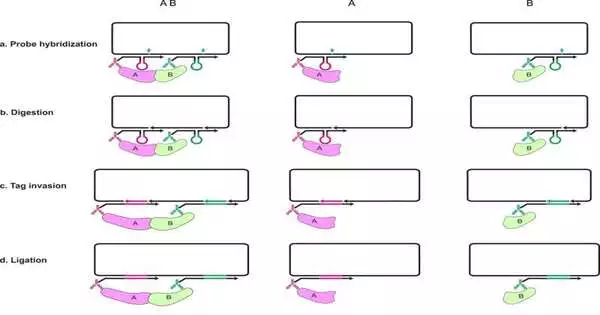
Schematic portrayal of the MolBoolean rule for the location of connecting and free proteins An and B. Nearness tests A (dark and red) and B (dark and green) hybridize to the circle after restricting their separate objective proteins An and B to the circle.”Bolts” means oligonucleotide extremity. The circle gets enzymatically scratched (the cyan-pointed stone shows the scratching position). c. The circle gets attacked by journalist labels (label A in red, label B in green). d Enzymatic ligation of the columnist’s labels to the circle follows. eRolling circle intensification (RCA) makes long concatemeric items (RCPs). RCPs are identified through fluorescently named tag-explicit location oligonucleotides. Nature Communications (2022) is credited.DOI: 10.1038/s41467-022-32395-w
“Our exploration means fostering devices to imagine processes inside cells and to increase, for instance, the information on what occurs in disease cells.” “Realizing the harmony among free and connecting proteins is significant while concentrating on cell flagging, and we accept that MolBoolean will be valued in many examination labs,” states Doroteya Raykova, analyst at the Department of Pharmaceutical Biosciences and first writer of the article.
MolBoolean was created by Ola Söderberg’s gathering at Uppsala University’s Faculty of Pharmacy. The work has been done as a team with scientists at the colleges of Porto and Uppsala, SciLifeLab and Atlas Antibodies.
More information: Doroteya Raykova et al, A method for Boolean analysis of protein interactions at a molecular level, Nature Communications (2022). DOI: 10.1038/s41467-022-32395-w
Journal information: Nature Communications