The most recent biological advances include gene editing. Prokaryotes (organisms without cell nuclei) gained resistance to foreign DNA thanks to the well-known CRISPR-Cas gene editing system. Researchers have been revealing the evolution of the CRISPR-Cas proteins from their precursors since the development of the CRISPR gene editing technology.
With this knowledge, they will be able to create new, compact genome editing tools for gene therapy. Professor Yokoi teaches at the University of Tokyo. The goal of Osamu Nureki’s team is to characterize the composition and purpose of the proteins used in genome editing. The team recently uncovered the 3D structure of a protein called TnpB, which is thought to be a precursor to the CRISPR-Cas12 enzyme. Nature published their research findings.
According to earlier research, the TnpB protein is thought to act as a pair of molecular scissors, severing DNA with the aid of a unique non-coding RNA called RNA. The exact mechanism of RNA-guided DNA cleavage and its evolutionary relationship with Cas12 enzymes, however, were unknown, which is what motivated Nureki Lab’s investigation. They had to reveal the protein structure, which was the first and most important step in understanding that.
“Our discoveries expand our knowledge of the development of TnpB proteins into CRISPR-Cas12 effectors and offer mechanistic insights into how TnpB functions.”
Ryoya Nakagawa, a graduate student and one of the first authors of the research paper.
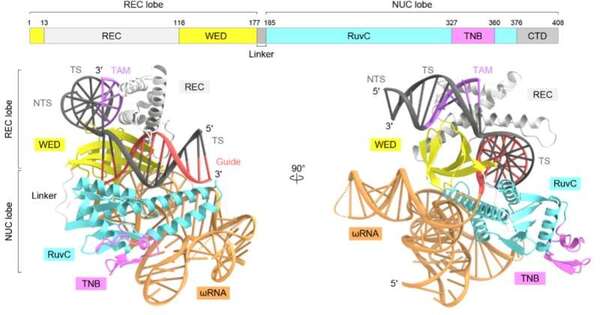
The scientists used cryo-electron microscopy and the protein TnpB from the bacterium Deinococcus radiodurans to determine the three-dimensional structure of the protein. The protein sample is cooled to -196°C using liquid nitrogen and exposed to electron beams during the cryo-electron microscopy process, which reveals the protein’s three-dimensional (3D) structure.
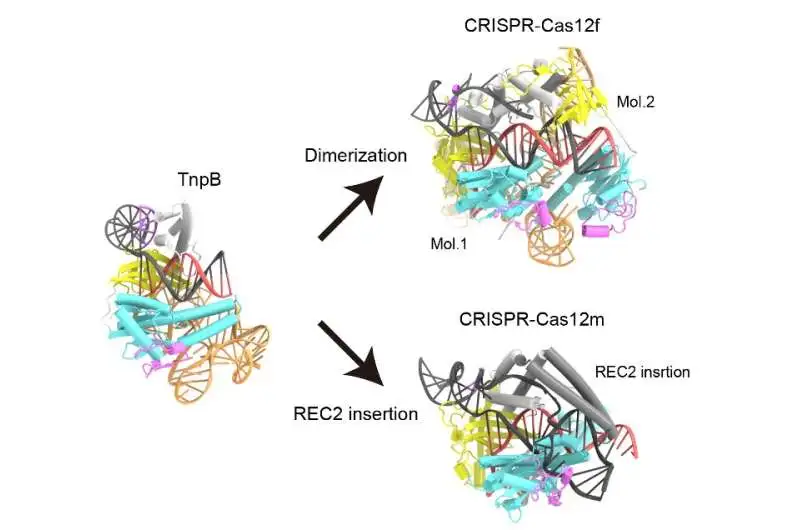
When TnpB and Cas12 enzymes with high sequence similarity to TnpB were compared structurally, it was discovered that Cas12 enzymes could engage in CRISPR-Cas adaptive immunity by either forming asymmetric dimers or using a variety of REC2 insertions to recognize longer target DNA sequences than TnpB. Credit: Nature (2023). DOI: 10.1038/s41586-023-05933-9
The research team discovered that the RNA in TnpB has a distinct pseudoknot shape resembling that of the Cas12 enzymes’ guide RNAs. The research also demonstrated how TnpB distinguishes the RNA and slashes the target DNA. Furthermore, they discovered two potential routes by which TnpB could have evolved into CRISPR-Cas12 enzymes by contrasting the structure of this protein with that of Cas12 enzymes.
Graduate student Ryoya Nakagawa, one of the study’s first authors, says that the findings “provide mechanistic insights into the function of TnpB and advance our understanding of the evolution from TnpB proteins to CRISPR-Cas12 effectors.”. He continued, “We will investigate future TnpB-based gene editing techniques’ potential applications.”.
More information: Ryoya Nakagawa et al, Cryo-EM structure of the transposon-associated TnpB enzyme, Nature (2023). DOI: 10.1038/s41586-023-05933-9