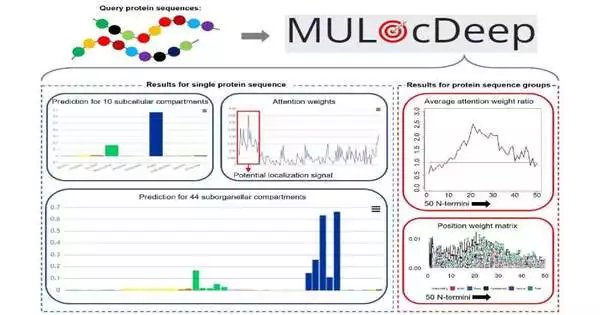
Researchers can unlock a wealth of biological information by predicting a protein’s location within a cell, which is essential for the development of future scientific discoveries related to the development of drugs and the treatment of diseases like epilepsy. This is due to the fact that proteins are the “workhorses” of the body and are mostly responsible for most cellular functions.
MULocDeep, the protein localization prediction model developed by Dong Xu and colleagues at the University of Missouri, which is a Curators Distinguished Professor in the Department of Electrical Engineering and Computer Science, was recently updated to include models for humans, animals, and plants. Xu and fellow MU researcher Jay Thelen, a professor of biochemistry, developed the model ten years ago to initially investigate proteins in mitochondria.
“Numerous organic revelations should be approved by tests, yet we don’t maintain that specialists should need to invest energy and cash directing a huge number of examinations to arrive,” Xu said. “Time is saved with a more targeted approach. Because we can assist them in designing more targeted experiments from which to advance their research more effectively, our tool serves as a resource that researchers can use to get to their discoveries more quickly.
“A more focused approach conserves time. Our platform helps researchers get to their discoveries faster by assisting them in designing more targeted experiments from which to advance their research more efficiently.”
Dong Xu, Curators Distinguished Professor in the Department of Electrical Engineering and Computer Science at the University of Missouri,
The model can assist researchers studying the mechanisms associated with irregular locations of proteins, or “mislocalization,” or where a protein goes to a different location than it is supposed to, by harnessing the power of artificial intelligence through a machine learning technique—training computers to make predictions using existing data. This abnormality is frequently linked to metabolic disorders, cancer, and neurological conditions.
“Mislocalization, which causes the protein to be unable to perform a function as expected because it either cannot go to a target or goes there inefficiently,” Xu stated, “Some diseases are caused by mislocalization.”
One more use of the group’s prescient model is helping with drug configuration by focusing on an inappropriately found protein and moving it to the right area, Xu said.
Xu intends to develop additional functionalities and improve the model’s accuracy in the future.
Xu stated, “We want to continue improving the model to determine whether a protein mutation could cause mislocalization, whether proteins are distributed in multiple cellular compartments, or how signal peptides can help predict localization more precisely.” Our tool may be of assistance to others in the creation of medical solutions, despite the fact that we do not provide any solutions for the development of drugs or treatments for various diseases. Science today is a huge enterprise. We can accomplish a lot of good for everyone by working together, even though different roles are played by different people.
Based on the biological and bioinformatics concepts used in the model, Xu and colleagues are currently developing a free online course for high school and college students. He anticipates that the course will be available later this year.
Xu and his colleagues also point to a conflict of interest: while the internet-based variant of MULocDeep is accessible for use by scholastic clients, an independent rendition is likewise accessible industrially through a permitting expense.
Nucleic Acids Research published a paper titled “MULocDeep web service for protein localization prediction and visualization at subcellular and suborganellar levels.” Co-creators are Yuexu Jiang, Lei Jiang, Chopparapu Sai Akhil, Duolin Wang, Ziyang Zhang, and Weinan Zhang at MU.
More information: Yuexu Jiang et al, MULocDeep web service for protein localization prediction and visualization at subcellular and suborganellar levels, Nucleic Acids Research (2023). DOI: 10.1093/nar/gkad374