Microorganisms, particularly bacteria, are skilled chemists who are capable of producing an impressive variety of natural products, which are chemical compounds. The microbes greatly benefit from the evolutionary advantages provided by these metabolites, which enable them to interact with one another or their environment and aid in defense against various threats. Many bacterial natural products have been utilized in medical treatments like antibiotics and drugs that fight cancer due to their numerous functions.
The microbial species alive today address just a small part of the huge variety of microorganisms that have possessed Earth over the past 3 billion years. There are exciting opportunities to recover some of their lost chemistry by investigating this microbial past.
Due to their poor preservation over time, it is virtually impossible to directly study these metabolites in archaeological samples. Notwithstanding, reproducing them using the hereditary outlines of long-dead organisms could be a way ahead.
Biochemists, anthropologists, and archaeogeneticists work together to investigate ancient microbes. By producing obscure substance compounds from the remade genomes of old microscopic organisms, our recently distributed research gives a proof of concept for the expected utilization of fossil microorganisms as a wellspring of new medications.
Remaking antiquated genomes
The phone apparatus that creates bacterial regular items is encoded in qualities that are ordinarily near each other, shaping what are called biosynthetic quality bunches. Such qualities are hard to identify and reproduce from antiquated DNA in light of the fact that exceptionally old hereditary material separates after some time, dividing into thousands or even great numbers of pieces. As a result, numerous tiny DNA fragments shorter than 50 nucleotides are assembled into a jumbled puzzle.
The genomes of millions of ancient bacteria are preserved in a single ancient tooth. Credit: Felix Wey/Werner Siemens Foundation
After sequencing billions of these ancient DNA fragments, we improved a bioinformatic technique known as de novo assembly to digitally arrange the fragments in stretches of up to 100,000 nucleotides—a 2,000-fold improvement. We were able to determine not only the number of genes that were present but also how they were arranged in the genome and how they differed from known bacterial genes—important information for understanding their evolutionary history and function.
This technique permitted us to investigate the genomes of microorganisms satisfying a long time ago, including species not known to exist today. Our findings push the earliest reconstructed microbial genomes back by more than 90,000 years.
A gene cluster that was shared by a high proportion of Neanderthals and anatomically modern humans living during the Middle and Upper Paleolithic, which lasted from 300,000 to 12,000 years ago, was found in the microbial genomes that we reconstructed using DNA extracted from ancient tooth tartar. This group of green sulfur bacteria, which are capable of photosynthesis, belonged to the bacterial genus Chlorobium and bore the molecular characteristics of extremely ancient DNA.
To enable the “modern” bacterium Pseudomonas protegens to produce the chemical compounds encoded in the ancient genes, we inserted a synthetic version of this gene cluster. Utilizing this strategy, we had the option to detach two obscure mixtures we named paleofuran An and B beforehand and decide their compound construction. Resynthesizing these atoms in the lab without any preparation affirmed their design and permitted us to deliver bigger amounts for additional examination.
By recreating these old mixtures, our discoveries show how archeological examples could act as new wellsprings for regular items.
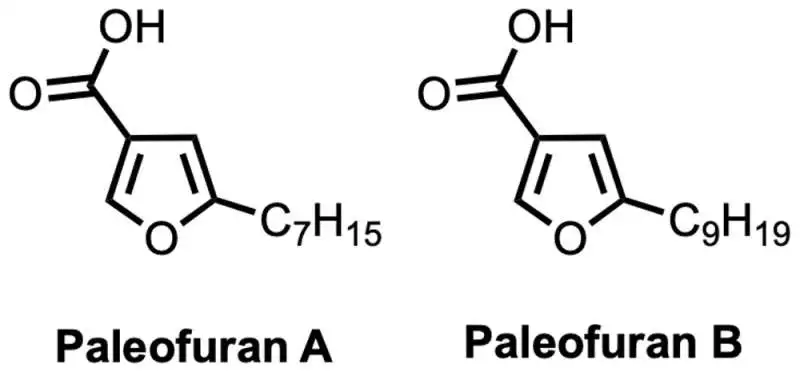
These paleofurans were produced from ancient microbial DNA. Credit: Pierre Stallforth, CC BY
Exploiting ancient natural resources
Microbes are always changing and adjusting to their surroundings. Microbes today probably produce different natural products than their ancestors did tens of thousands of years ago because they live in different environments.
As late as 25,000 years ago, the Earth went through a significant environmental shift as it changed from the colder and more unstable Pleistocene Age to the hotter and more calm Holocene Age. During this time, people began to live outside of caves and began increasingly experimenting with food production, which led to significant changes in human lifestyles. These progressions brought them into contact with various microorganisms through agribusiness, creature farming, and their new assembled conditions. Concentrating on Pleistocene-period microorganisms might yield bits of knowledge into bacterial species and biosynthetic qualities that are not related to people today, and maybe even organisms that have become extinct.
While the amount of information gathered by researchers on natural organic entities has dramatically expanded throughout the course of recent years, the quantity of new anti-toxins has deteriorated. When bacteria are able to evade existing antibiotic treatments faster than researchers can develop new ones, this becomes especially problematic.
Scientists can discover the hidden diversity of natural products that would have been lost over time by reconstructing microbial genomes from archaeological samples. This increases the number of potential sources from which they can discover new drugs.
Scaling up ancient molecules Our research has demonstrated that ancient natural products can be accessed. We now need to simplify our approach to make it less labor-intensive in order to gain access to the extensive variety of chemical compounds that are encoded in ancient DNA.
In order to more quickly and accurately identify biosynthetic genes in ancient DNA, we are currently improving and automating our method. In addition, to finish the time-consuming pipetting and bacterial cultivation steps in our methods, we are implementing robotic liquid handling systems. Our objective is to scale up the process so that we can turn a lot of information about ancient microbes into new therapeutics.
In spite of the fact that we can reproduce old particles, their organic and environmental jobs are hard to unravel. Since the microbes that initially created these mixtures never again exist, we can’t culture or hereditarily control them. Future research will need to use bacteria that are similar to those found today. It remains to be determined whether these compounds’ functions have remained unchanged in modern relatives of ancient microbes. Even though we don’t know what these compounds originally did for ancient microbes, they could still be used to treat modern diseases.
By providing a brand-new time axis for antibiotic discovery, our ultimate goal is to provide fresh insight into microbial evolution and combat the current antibiotic shortage.
Provided by The Conversation





