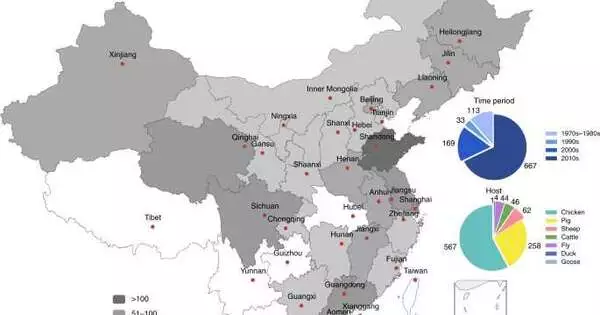
A multinational team of experts discovered that there is a rising resistance to antibiotics used in China through a genetic examination of bacteria in livestock. The group reveals their whole-genome study of Escherichia coli samples gathered from a vast number of pigs, chickens, cattle, and sheep farmed in China between 1970 and 2019. Their paper was published in the journal Nature Food. They searched for genes that give antibiotic resistance to a variety of regularly used antibiotics. They searched for genes that give antibiotic resistance to a variety of regularly used antibiotics. They also explain why they feel their findings demonstrate that China and other countries must develop innovative strategies to combat bacterial illnesses in both humans and animals. Claire Heffernan of the London International Development Centre released a News & Views essay in the same journal issue describing the history of antibiotic usage in agriculture and the team’s work on this new initiative.
An international team of researchers has found, through genetic testing of bacteria in livestock, that there is growing resistance to antibiotics used in China. In her article published in the journal health food, the group describes their whole genome analysis of Escherichia coli samples collected from large numbers of pigs, chickens, cattle and sheep reared in China between 1970 and 2019.
the journal health food
Scientists all across the world have been warning about rising antibiotic resistance in the treatment of human diseases in recent years. However, animals are also a part of the problem. As Heffernan points out, agriculture, particularly cattle, consumes around two-thirds of all antibiotics provided each year. The researchers asked what effect such use would have on the development of resistance to antibiotics used to prevent or cure bacterial illnesses such as E. coli in this new endeavor.
To find out, the researchers collected 986 samples of bacteria from animals collected by Chinese entities during the last half-century and submitted them to whole-genome analysis, specifically looking for alterations in genes that might confer resistance to antibacterial drugs. In all of the livestock investigated, they discovered signs of a slowly building resistance to drugs against E. coli. The overall increase is substantial, implying that antibiotics will soon be ineffective in protecting animals. The researchers also evaluated the antibiotic susceptibility of modern E. coli strains and discovered that they were considerably more resistant than bacteria from the 1970s. They also point out that because the same antibiotics are used to treat the same germs, the rapid growth in resistance also applies to human use.
Bacterial resistance is a difficult scientific problem. To address this issue, we must first thoroughly grasp the driving elements and methods. This research examines the factors that influence the acquisition of bacterial resistance in the context of cattle husbandry. In the meantime, the resistance mechanism is discussed. “Survival of the fittest” is the result of bacterial pathogens’ genetic flexibility, which causes specialized responses such as creating adaptive mutations, acquiring genetic material, or modifying gene expression. To a considerable extent, bacterial populations acquire resistance genes as a result of antibiotic-induced selective pressure. Mobile resistance genes, on the other hand, may be co-selected by other existing compounds (such as heavy metals and biocides) in the absence of direct selection pressure from antibiotics. This is because the resistance determinants of some of these drugs can co-locate the same mobile genetic elements as antibiotic resistance genes. Furthermore, environmental variables contribute to the acquisition of resistance genes. In this section, we will discuss some of the most important steps that should be taken to reduce the danger of antibiotic resistance. We urge relevant governments and organizations around the world to develop and improve antibiotic resistance monitoring strategies, boost supervision, increase international collaboration and exchange, and prevent the formation and spread of drug-resistant strains.
The formation and dissemination of antibiotic resistance in bacterial pathogens is a natural reaction to the selection pressure of antibiotics (Witte et al., 1999). According to the most recent estimates, total global sales of antimicrobial drugs for livestock were 93,309 tons in 2017, with sales expected to rise 11.5 percent to 1,04,079 tons by 2030.(Tiseo et al., 2020). Antibiotics are used in animals for three major reasons: to treat pathogen infections (therapeutic usage), to avoid animal disease outbreaks (prevention), and to promote growth (Gould, 2016; Roth et al., 2019). Antibiotic-resistant bacteria linked with animals are easily transmitted to people and spread in the environment via animal excrement (Van Boeckel et al., 2015; Manyi-Loh et al., 2018). As a result, when bacterial infections affect animals, most governments support antibiotic treatment. This medication can successfully prevent disease spread and reduce pathogen excretion in animals. In many developing nations and a few industrialized countries, antibiotics are used on animals as a preventive approach to prevent illness outbreaks, even when the animals are healthy and there are no known infectious diseases in the feeding environment (Prescott, 2017; Collineau et al., 2018).
Finally, the use of antibiotics as growth promoters (AGPs) in developing countries’ animal husbandry has been a source of contention. In the review (Sharma et al., 2016), for example, the research described the types of antibiotics registered as feed additives for cattle (Hobman and Crossman, 2015) and poultry in the United States, Canada, the European Union, and Australia to promote healthy growth. When antibiotics are fed to farm animals at low or sub-dose quantities, 75–90% of the antibiotics are lost to the environment in the form of urine and feces (Lekshmi et al., 2017). However, the presence of sub-therapeutic antibiotics in some tissues or organs (such as the intestines) causes non-specific bacterial mutations, which may encourage the development of bacterial resistance (Cogliani et al., 2011). Furthermore, the low amounts of heavy metals in the intestines and feces frequently come into contact with microorganisms. Heavy metals are typically incorporated into feed as nutritional supplements since they may be required elements to support certain physiological functions of the animal body (Hejna et al., 2018). On the other hand, some heavy metals, with no known biological activities, are regarded as contaminants. Under the effect of heavy metals, microorganisms have acquired antibiotic resistance in recent years (especially copper and zinc). Heavy metals, unlike antibiotics, are not biodegradable in many settings at relatively low concentrations (lower than their lowest inhibitory concentration), and their effects are extensive and long-lasting (Luo et al., 2014; Waseem et al., 2014; Gumpu et al., 2015; Zhao et al., 2015). Overall, metals can exert long-term selective pressure on microbes. Some evidence suggests a link between heavy metal resistance genes and antibiotic and disinfectant resistance genes in food-borne Salmonella (Yang et al., 2016; Zhang et al., 2016; Deng et al., 2018). Because of their disinfectant properties and ability to cure wounds, biocides are widely utilized in livestock husbandry. Biocides employed in a variety of applications eventually make their way into the environment. Fungicides, even at low doses, clearly impose selective pressure on microorganisms (Ons et al., 2020). The mechanisms that lead to resistant bacterial populations as a result of antibiotic use, heavy metal use, and biocide exposure have all received a lot of study. Furthermore, relevant research has demonstrated that when bacteria are exposed to these non-antibiotic drugs, they can be induced or selected to exhibit adaptation, diminishing sensitivity to one or more antibiotics (Wales and Davies, 2015). For example, after being exposed to phenolic fungicides or sub-inhibitory triclosan doses, the prevalence of antibiotic resistance mutations in Salmonella increases (Randall et al., 2004).
Pathogens will also be spread in other ways, such as through the food chain from animals to people (Gupta et al., 2020; Lei et al., 2020). Furthermore, bacteria can acquire exogenous resistance genes from other environmental sources, such as flies (Zhao et al., 2019). Figure 1 depicts the sources of selective pressure and the paths of drug resistance transfer. Vertical gene transfer (VGT), a cell development mechanism (Seoane et al., 2011), and horizontal gene transfer (HGT), the transfer of DNA between genera by mobile plasmids (Li et al., 2019a), coexist in natural habitats (Qiu et al., 2018). Under the strain of long-term selection, the indivisible link between humans, animals, the environment, and food may play an essential role in causing drug resistance and causing vertical or horizontal transfer of drug-resistant characteristics.