A microbial standoff beneath the prickly spines of European hedgehogs may have bred a dangerous drug-resistant pathogen long before the era of antibiotic use in humans. Antibiotic use, without a doubt, accelerates drug resistance in bacteria that colonize humans, according to Jesper Larsen, a veterinarian at Statens Serum Institut in Copenhagen. However, he claims that these microbes had to acquire the resistance genes from somewhere, and scientists aren’t sure where the majority of these genes come from.
Now, for one type of methicillin-resistant Staphylococcus aureus, or MRSA, Larsen, and colleagues have tracked its evolution to hedgehogs hundreds of years ago. On the skin of these critters, a fungus that produces natural antibiotics may have created the environment for drug resistance to evolve in the bacteria, the researchers report in Nature.
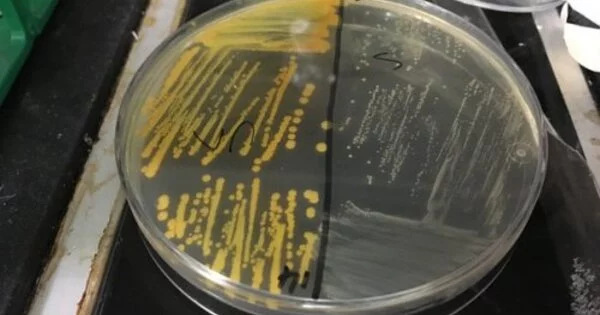
MRSA, one of the most common drug-resistant pathogens, infects hundreds of thousands of people each year around the world, and these infections can be difficult to treat. The new study focuses on a subset of MRSA that causes only a small percentage of human cases. Years ago, biologist Sophie Rasmussen, who was involved in the new research and is now at the University of Oxford, approached Larsen’s team about sampling a freezer full of dead hedgehogs by chance. MRSA was found in 61% of the animals collected in Denmark. “We discovered an extremely high prevalence in hedgehogs,” Larsen says, implying that the animals served as a reservoir for the drug-resistant superbug.
There is no doubt that our use of antibiotics is the main driver of resistance in human pathogens. This is a very special case where we can simply trace it back to its origin.
Anders Larsen
In the new work, the scientists surveyed hedgehogs (Erinaceus europaeus and Erinaceus roumanicus) from 10 European countries and New Zealand. Workers at wildlife rescue centers swabbed the noses, skin, and feet of 276 animals. MRSA was prevalent in hedgehogs in the United Kingdom, Scandinavia, and the Czech Republic.
Analyzing the S. aureus, the team found 16 strains of mecC-MRSA, named after the gene that confers resistance, and mapped the evolutionary relationships between them by comparing mutations across their genetic instruction manuals, or genomes. From the analysis, the team inferred that the three oldest lineages emerged 130 to 200 years ago in hedgehog populations, periodically infecting people and cattle long before penicillin hit the market in the 1940s. Hedgehogs may be the source of nine out of the 16 lineages, the researchers report.
“There is no doubt that our use of antibiotics is the main driver of resistance in human pathogens,” says Anders Larsen, a microbiologist at Statens Serum Institut who was also a member of the team. “This is a very special case where we can simply trace it back to an origin.”
However, this does not explain how the hedgehogs’ S. aureus developed resistance. A clue came from a 1960s research study on Trichophyton erinacei, a fungus that causes “hedgehog ringworm” in humans. T. erinacei on hedgehog skin killed some S. aureus but not others that were penicillin resistant. Growing T. erinacei in the lab, the researchers discovered two penicillin-like antibiotics produced by the fungi.
This finding suggests that hedgehogs are a MRSA reservoir because “they’re living cheek by jowl with organisms that are producing penicillin,” says Gerry Wright, a biochemist at McMaster University in Hamilton, Canada, who was not involved with the study.
Wright describes the fungi as “living in a bad neighborhood.” They must compete for resources and a place to colonize on the host with other microbes such as S. aureus, and “they must work out this arrangement where they can protect themselves.”
Wright believes that thinking about antibiotic resistance without considering environmental implications is impossible. According to him, the evolution of resistance is a gradual process shaped by natural selection. Wright’s research has shown that antibiotic resistance has ancient origins in areas that have escaped human influence. This evolution has primarily been sought in the soil microbial community, or microbiome. However, he claims that animal microbiomes provide another potential source for resistance genes as well as sources of new antibiotics.
The history of antibiotics over the last century has been a cycle of new drug discoveries followed by microbial resistance to those drugs. That should come as no surprise, according to Wright. “Because antibiotics have been around for billions of years, and resistance has been around for billions of years,” he explains. He claims that if scientists do not better understand where resistance comes from, even if new drugs are discovered, we will be playing catch-up.