Exact control of quality articulation—gguaranteeing that cells make the right parts in the perfect sum and with impeccable timing—iis fundamental for all creatures to appropriately work. Cells should have control over how DNA-encoded qualities are converted into RNA particles that can perform cell functions on their own or be processed into proteins.
One way quality articulation is controlled is by stopping “record”—the interaction by which RNA is integrated from its DNA format by a chemical called RNA polymerase. Scientists have now discovered the component of record-stopping in specific microorganisms using cryo-electron microscopy (cryo-EM), allowing them to choose to nuclear scale the designs of the RNA polymerase before, during, and soon after a delay in RNA creation.Explaining the system of stopping records is significant to figuring out essential cell capabilities.
One of the vital parts of record stopping in the microbes is a protein factor called NusG, which is conserved across creatures, including people, to such an extent that the stopping component uncovered by this study might be comprehensively useful for figuring out quality guidelines in all organic entities on the planet. The knowledge could likewise be utilized to recognize new enemies of bacterial specialists that objectively hinder record stopping, subsequently upsetting appropriate quality articulation and cell capability.
“In order to function properly, cells must carefully control gene expression to guarantee that the relevant proteins and functional RNAs are produced in the appropriate amount and at the appropriate time.”
Kastuhiko Murakami, professor of biochemistry and molecular biology at Penn State
A paper depicting the exploration by a group of Penn State researchers appears online Feb. 6 in the journal Procedures of the Public Foundation of Sciences.
“To work appropriately, cells should unequivocally control quality articulation to guarantee that they are making the chosen proteins and useful RNAs in the suitable sum and at the proper time,” said Kastuhiko Murakami, teacher of natural chemistry and sub-atomic science at Penn State and one of the heads of the examination group.
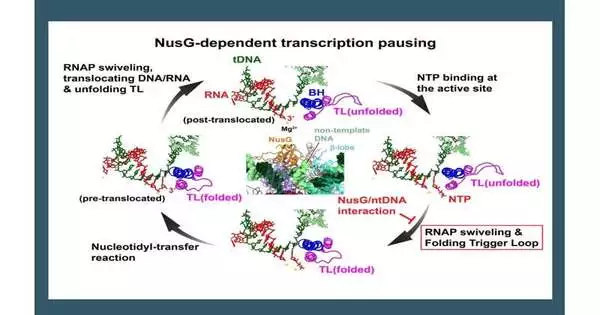
“Quality articulation can be controlled in more ways than one in any event, when RNA polymerase is effectively making RNA, for example, by stopping the recording.” Here, we utilized cryo-EM structure assurance and construction-based biochemical measures to recognize the collaborations among RNA polymerase, DNA, and NusG, as well as the primary changes of RNA polymerase that happen when RNA polymerase stops during recording.
RNA polymerase is a sub-atomic machine made out of a few useful spaces. It likewise interfaces with various other protein co-factors that assist with directing the timing and amount of RNA created. Work in the lab of Paul Babitzke, Stanley Individual Teacher of Sub-atomic Science at Penn State and head of the examination group, had shown that one of those co-factors, the protein called NusG, was pivotal for stopping the record.
“We showed tentatively that NusG assumed a significant role in stopping the record by perceiving a particular short DNA succession theme,” said Babitzke.
“We saw more than 1600 of these grouping themes across the genome of the microorganisms, Bacillus subtilis, that are engaged in NusG-subordinate stopping of record. DNA is split in two, and only one strand, known as the layout strand, is deciphered into RNA.Curiously, the acknowledgement theme for NusG is situated on the non-layout strand. To understand how NusG collaborating with non-layout DNA could cause a stop, we needed to look at what was going on at the complex level.”Cryo-EM has permitted us to do precisely that.”
Remaking of a video showing the nucleotide expansion cycle of RNA polymerase, NusG-subordinate stopping, and breaking from the stopped record.Credit: Murakami Research Center
The exploration group distinguished DNA and RNA successions that make records stop within the sight of NusG and afterward reconstituted the record complex utilizing DNA, RNA, and NusG along with RNA polymerase. The samples were frozen and then used for underlying investigations by cryo-EM at the Penn State Huck Organizations of the Existence Sciences’ cutting-edge cryo-electron microscopy office.
In the design of the complicated stopped record, the group uncovered that NusG communicates with the non-format DNA strand by embedding it into a thin pit between NusG and a space for RNA polymerase called the beta curve. In any case, this cooperation happens far away from the dynamic site of RNA polymerase, so how can it make records stop? To answer this question, the investigation team used one-of-a-kind data obtained from the cryo-EM underlying review.
“At the point when we freeze the example, the reconstituted record edifices can each be at somewhat various stages in the record cycle,” said Rishi K. Vishwakarma, collaborator and research teacher of organic chemistry and sub-atomic science at Penn State and the main creator of the paper. “With cryo-EM, we can utilize information from the single example to decide on a progression of designs showing unmistakable phases of record stopping, then orchestrate these designs consecutively.” We utilized them to reproduce a film of record edifices to see what befalls the record complex when recording stops, similar to how a flipbook can address development from a progression of still pictures.
“It required an investment to explain how the communication of NusG and the non-format DNA stops recording,” said Murakami.
“One evening, while watching a film of not entirely settled in this work, I noticed that an enormous conformational change of RNA polymerase—the turn module revolution—is linked to set off circle collapsing, a fundamental conformational change of RNA polymerase for RNA combination. At the point when the non-format DNA is caught between NusG and the RNA polymerase beta curve, it prevents the pivot of the turn module, accordingly slowing down trigger circle collapse and RNA synthesis, similar to stalling something out in one of the cog wheels of a motor that will affect different pieces of the motor and stop the vehicle. “I was extremely excited for this finding and couldn’t quit watching the film as my thoughts hardened.”
Because record stopping is a transient component for halting RNA blend, the group also examined the primary changes of the record puzzle as it moved away from stopping by constructing an example containing the stopped record intricate and the RNA polymerase substrate (NTP) and determining cryo-EM structures.
Yet again in the record complex, which had gotten away from the delay, the non-format DNA was not generally caught between NusG and the RNA polymerase beta curve, which permitted the turn module to pivot to such an extent that the trigger circle could overlap appropriately.
“Since the greater part of the parts of this cycle are moderated by everything from microbes to people, we are keen on knowing whether the instrument is likewise monitored,” said Murakami.
“We are working with other cell RNA polymerases, for example, from archaea and eukaryotes. We were lucky to get an eukaryotic RNA polymerase disconnected from Drosophila (natural product flies) from David Gilmour at Penn State, who has been concentrating on record-breaking in eukaryotes for a really long time. “We will determine how to search for stopped RNA polymerase structures in archaea and eukaryotic life forms.”
More information: Rishi K. Vishwakarma et al, Allosteric mechanism of transcription inhibition by NusG-dependent pausing of RNA polymerase, Proceedings of the National Academy of Sciences (2023). DOI: 10.1073/pnas.2218516120