Although it is in the near future, the idea of storing data in DNA sounds like something out of a science fiction novel. Within five to ten years, Professor Tom de Greef anticipates that the first DNA data center will be operational. A hard drive will not store data as zeros and ones but rather as the base pairs of DNA: CG and AT. Such a server farm would appear as a lab, commonly more modest than the ones today.
De Greef already sees everything. In one piece of the structure, new records will be encoded through DNA combinations. Large fields of capsules, each containing a file, will be in another section. A capsule will be removed, its contents read, and then returned by a robotic arm.
The topic at hand is synthetic DNA. In the lab, bases are kept together in a specific order to frame artificially created strands of DNA. DNA can then be used to store photos and files that are currently stored in data centers. The method can only be used for archival storage at this time. You want to consult the DNA files as little as possible due to the high cost of reading stored data.
Large, energy-guzzling data centers made obsolete
Data storage in DNA has numerous advantages over large, energy-intensive data centers. A DNA record can be put away considerably more minimalistically, for example, and the life expectancy of the information is likewise ordinarily longer. Most importantly, however, this new technology eliminates large, energy-intensive data centers. Furthermore, this is frantically required, cautions De Greef, “on the grounds that in three years, we will create such a lot of information overall that we will not have the option to store half of it.”
De Greef and Ph.D. student Bas Bögels, Microsoft, and a group of university partners have developed a new method to scale the data storage innovation using synthetic DNA. The outcomes have been distributed today in the journal Nature Nanotechnology. De Greef is a professor at Radboud University and teaches at the Institute for Complex Molecular Systems (ICMS) and the Department of Biomedical Engineering at TU Eindhoven.
Adaptable
Involving strands of DNA for information capacity arose during the 1980s but was unreasonably troublesome and costly at that point. When DNA synthesis began to gain momentum three decades later, it became technically feasible. In 2011, Harvard Medical School geneticist George Church elaborated on the concept. Since then, the cost of data synthesis and reading has decreased dramatically, bringing the technology to market.
De Greef and his group have focused primarily on reading the stored data in recent years. This is currently the most significant issue with this new method. The “random access” PCR method that is currently being used for this is extremely error-prone. You can hence just read each record in turn, and, likewise, the information quality breaks down a lot each time you read a document. Not precisely adaptable.
How does it work? By adding a primer containing the desired DNA code, PCR (polymerase chain reaction) generates millions of copies of the required piece of DNA. Crown tests in the lab, for instance, depend on this: When copied so many times, even a tiny amount of your nose’s coronavirus can be detected. However, you need multiple primer pairs working simultaneously to read multiple files simultaneously. The process of copying suffers greatly as a result of this.
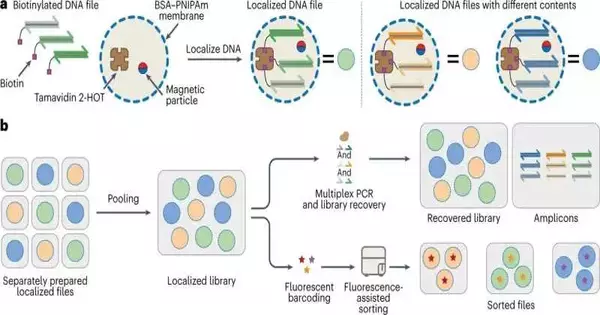
Each container contains one record.
This is where the containers become possibly the most important factor. The team led by De Greef created a microcapsule made of proteins and a polymer, anchored with one file per capsule. “These capsules have thermal properties that we can use to our advantage,” states De Greef. At temperatures above 50 degrees Celsius, the capsules self-seal, allowing each capsule to undergo its own PCR procedure. There is then little room for error. This is known as “thermo-confined PCR” by De Greef. It has so far been able to read 25 files simultaneously in the lab without making a lot of mistakes.
After that, if you lower the temperature once more, the copies fall out of the capsule, but the anchored original stays, so the quality of your original file stays the same. De Greef says, “After three reads, we currently stand at a loss of 0.3 percent, compared to 35% with the existing method.”
Fluorescence-searchable, but that’s not all. Additionally, De Greef has improved the data library’s searchability. Each capsule has a unique color and a fluorescent label for each file. A gadget can then perceive the varieties and separate them from each other. This brings us back to the fictitious robotic arm that was introduced at the beginning of the story. In the future, it will precisely select the desired file from the pool of capsules.
The issue of reading the data is solved by this. According to de Greef, “Presently, it’s simply an issue of holding on until the expenses of DNA combination fall further. After that, the method will be ready to be used.” He hopes that the Netherlands will soon be able to open its first DNA data center, which would be a first in the world.
More information: Yuan-Jyue Chen, DNA storage in thermoresponsive microcapsules for repeated random multiplexed data access, Nature Nanotechnology (2023). DOI: 10.1038/s41565-023-01377-4. www.nature.com/articles/s41565-023-01377-4