Analysts in Japan have found that the nearby DNA movement within human cells stays consistent all the way through interphase, where the cell develops and imitates its DNA for cell division. The review proposes that this consistent state of DNA movement permits cells to lead housekeeping errands under comparable conditions during interphase.
The group, led by Professor Kazuhiro Maeshima of the National Institute of Genetics, ROIS, published their discoveries on June 3 in Science Advances.
To fit inside the core of the cell, DNA is coordinated into chromatin, in which the strands of DNA are folded over gatherings of histone proteins, similar to string around a spool, to shape structures known as nucleosomes. Nucleosomes can then be collapsed up into significantly more conservative designs and structure chromatin. Past examination shows that chromatin is constantly influenced in living cells.
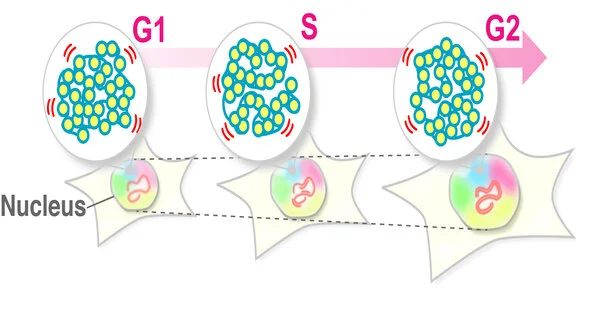
As the cell cycle advances (in particular the G1, S, and G2 stages), where genome DNA pairs and the core expands, the atomic climate encompassing chromatin radically changes. Maeshima and partners at the National Institute of Genetics in Mishima, Japan, built this inquiry: how does chromatin conduct change during interphase?
Maeshima’s group utilized a high-goal light microscopy strategy to take a gander at the way of behaving of individual nucleosomes inside living cells for an extremely brief time frame, roughly one second.
Maeshima and colleagues have uncovered that the nearby chromatin movement stays consistent all through interphase, despite the fact that genome DNA is multiplied by DNA replication and the core develops. The analysts, likewise, have shown that atomic development without replication didn’t influence the consistent state movement of chromatin. Along these lines, neighborhood chromatin movement is autonomous of such atomic changes during interphase.
“This is a significant finding in light of the fact that the consistent state movement permits cells to direct their schedules, like RNA replicating and DNA replication, under comparable atomic conditions,” the principal creator, Shiori Iida, said. “Nearby chromatin movement can oversee genomic DNA openness for target looking or selecting a piece of hardware.” “The consistent state movement of chromatin gives a vigorous cell framework in which DNA capacities are unaffected by different atomic changes.”
“Cells can momentarily change the chromatin movement from the consistent state to play out their impromptu positions because of DNA damage, among numerous different assignments,” Maeshima said. He and his group plan to additionally investigate how DNA movement is directed, which proteins are associated with the guideline interaction, and how DNA acts during cell division. “Our definitive objective is to comprehend how human genomic DNA inside the cell acts to peruse out hereditary data in it,” Maeshima said.