Long periods of work by formative scholars from Trinity College Dublin, working with partners at the University of Edinburgh’s MRC Human Genetics Unit, has yielded an asset that will assist scientists with surveying how key qualities control the separation of tissues and organs of creating incipient organisms.
The cooperative group, which has recently distributed the work in the diary, Development, examined these key qualities, their correspondences, and their effects in the 3D space of the mouse undeveloped organism across various progressive phases.
We plunked down with Professor Paula Murphy, from Trinity’s School of Natural Sciences, to figure out more about the examination and its more extensive ramifications.
What leap forward has this work taken?
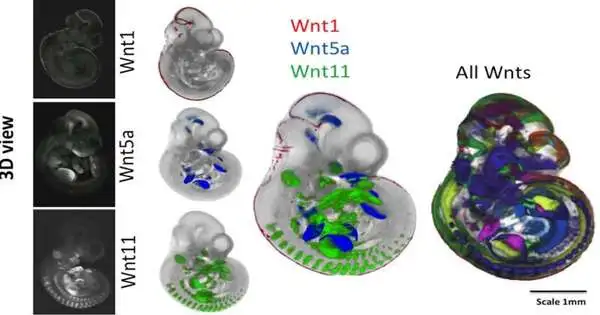
Certain qualities and quality organizations play an important role in determining how an embryonic organism develops, how different cell types (muscle, bone, and neuron) structure, and where organs appear. For years and years at this point, utilizing sub-atomic science methods, we could find where and when one of these “WNT qualities” is turned on, assisting us with understanding how it works in directing early stage advancement.
The method would need to be performed for every quality separately to get a ‘preview’ of where it is communicated in 3D undeveloped organism space at a specific time. What we have figured out how to accomplish here is the troublesome errand of planning these spatial information previews for some qualities, coordinating and looking at where every quality is turned on inside the creating incipient organism. Fostering the devices to have the option to do this was a significant accomplishment for our computational science partners at the Edinburgh Mouse Atlas project.
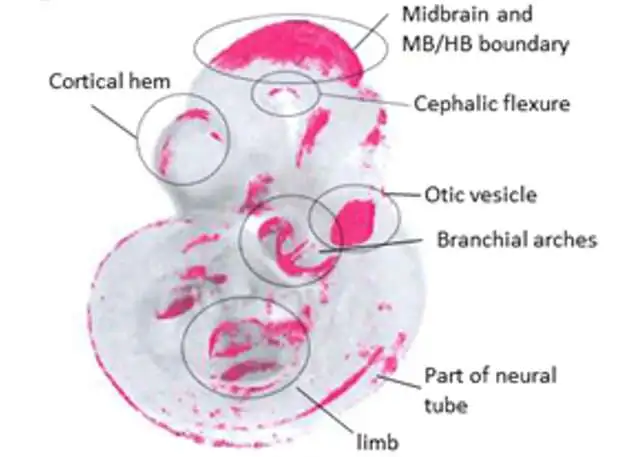
The outer perspective on a 10.5-day old mouse undeveloped organism shows the “problem areas” where various flagging qualities are co-communicated in red.
Is it true that you were amazed by anything you found?
This approach permitted us to make various revelations that could never have been conceivable without coordinating (planning) the examples in general. For instance, we could uncover where the pathway is quiet since one or other of the parts isn’t enacted.
Likely the most clever finding was that there are “problem areas” in the undeveloped organism where a great large number of these qualities are turned on together. Some of these problem areas are known to be the areas of significant gatherings of cells that guide improvement, yet some are already obscure and uncover already undetectable flagging places in the undeveloped organism.
What other research question(s) might prospective scholars consider at this time?
Going with this distribution, we have made each of the crude and planned information openly accessible, as well as the apparatuses for imagining them, so any formative scholar can pose questions about this cell correspondence framework in any piece of the undeveloped organism. Associates interested in kidney advancement or mental health, for example, can delve deeply into the framework parts activated in that part of the developing organism.
We and others can now use this data to ask questions about how the framework has evolved.For instance, there are 19 very much like qualities in a mouse and a human being. These various qualities have emerged through quality duplication an extremely long time ago. We can now inspect how the qualities have veered, each taking on novel capabilities during the improvement of a complex multicellular incipient organism.
What are the next steps for your group?
We are right now doing a centered examination of the coordinated informational index in the creating appendage buds to see how the cell correspondence framework works in the appendage. We also anticipate seeing how the information will be used by partners with various professional interests; for greater comprehension, we may interpret each developing framework.
More information: Paula Murphy et al, Integrated analysis of Wnt signalling system component gene expression, Development (2022). DOI: 10.1242/dev.200312
Journal information: Development