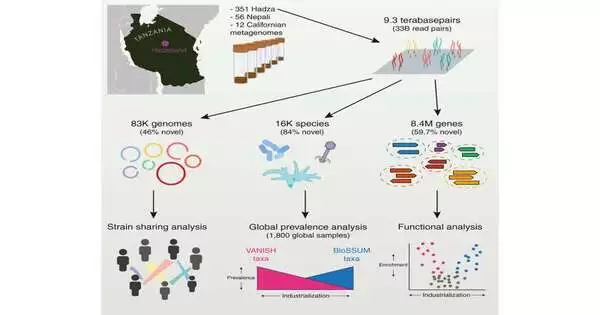
Ultra-deep metagenomic sequencing of 351 feces samples from the Hadza hunter-gatherers of Tanzania was carried out as part of a genomic study that was led by scientists at Stanford University in California. The team describes how they obtained the most comprehensive set of gut microbiome sequencing data from a hunter-gatherer population and identified it in their paper, “Ultra-deep sequencing of Hadza hunter-gatherers recovers vanishing gut microbes,” which was published in Cell.
Depending on local diet and environmental contact, microbiome composition can vary significantly across the globe. Microbiome research is heavily influenced by Western populations with low microbiome diversity. Consumption of highly processed foods, an increase in the use of antibiotics, the rate of cesarean sections, sanitation levels, and a decrease in physical contact with animals and soil are thought to be the root causes of the low diversity.
By looking at populations that have not been industrialized, such as hunter-gatherer populations, we can see that there is a causal connection. These populations also have extremely diverse microbiomes. It can also be seen as a shift toward less diversity in the microbiome among immigrants from non-industrialized regions to industrialized Western cultures.
“A significant challenge will be to characterize the impact of these microbes on human physiology and determine whether the absence or presence of species and functions is beneficial or detrimental to human health in different contexts.”
The high diversity of the modern non-industrialized microbiome resembles the ancient microbiome more than the industrialized one, according to previous research on ancient human coprolites.
Given that the gut microbiota plays a crucial role in human health and that these human-associated microbial lineages have been passed down through human generations over long periods of evolutionary time, it follows that the absence of these microbes’ functions in the industrialized microbiome is a loss.
The Hadza people in this study live in camps of five to thirty people each in the central Rift Valley of Tanzania. About every four months, people move around the camp. They fundamentally drink from springs and streams and have an eating regimen that incorporates scrounged tubers, berries, honey, and chased creatures.
Stool samples from 167 Hadza people were used for metagenomic sequencing. The Unified Human Gastrointestinal Protein Database does not include more than half (59.7%) of the 8.4 million protein families that are found in Hadza gut microbes. The global list of microbiota that are known to exist rises by approximately 24% due to the inclusion of bacterial and viral species in the microbiome.
The team compared the findings to adult Californians for a local comparison and discovered that, whereas the average Hadza adult gut microbiome contains 730 species, the Californian gut microbiome only contains 277 species.
“An important challenge is to characterize the impact of these microbes on human physiology and determine in which contexts the absence or presence of species and functions is beneficial or detrimental to human health,” the authors declare.
It will be fascinating to learn how Western metabolisms are affected by a lack of diversity, as well as which diseases, if any, the Hadza populations are avoiding. Although it’s possible that the reasons no longer apply, it’s unlikely that the missing microbiota from industrialized populations was there without a reason.
Future researchers will have access to an extremely deep sequenced data set and a wealth of new mysteries to solve as a result of the current effort.
More information: Matthew M. Carter et al, Ultra-deep sequencing of Hadza hunter-gatherers recovers vanishing gut microbes, Cell (2023). DOI: 10.1016/j.cell.2023.05.046