Scientists all over the world are looking for ways to combat bacteria that can’t be killed by antibiotics to avoid a global health crisis.
A promising objective for better-than-ever anti-infection agents are riboswitches, little stretches of RNA that direct a cycle important for the development of proteins by the bacterial cell. Riboswitches are tracked down only in microorganisms and could be focused on with anti-infection agents, so creatures or people are unaffected. Researchers may be able to develop drugs that disrupt the cellular machinery that produces necessary proteins once they have a complete understanding of how riboswitches function.
Using a combination of biochemistry, structural biology, and computational modeling, researchers at the University of Michigan’s Life Sciences Institute and the Department of Chemistry have now revealed how a particular riboswitch regulates its own synthesis.
Transcription is the first step in making a protein from the genetic code. The genetic information in DNA is copied into a strand of RNA by the enzyme RNA polymerase (or RNAP), which moves along the DNA. RNAP will go through a number of “pauses” as it waits for the cell to give it more instructions during this process. Scientists have been trying for a long time to figure out how this pause and restart happens, but antibiotics could use this as a perfect target.
“This work is an excellent example of the University of Michigan’s scientific strength. Three labs with disparate expertise were able to develop a multidisciplinary collaboration, which resulted in a significant and original finding.”
Professor Melanie Ohi
Using a structural biology technique known as single particle cryo-electron microscopy (cryo-EM), the team, which was led by chemistry professor Nils Walter and collaborated with the labs of LSI professor Melanie Ohi and former U-M scientist Aaron Frank, was able to see for the first time how this transcriptional regulation occurs. Nature Structural & Molecular Biology has published their findings.
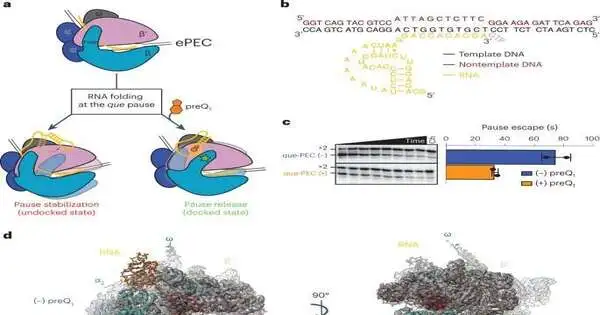
The Walter lab took a gander at a specific riboswitch that ties a particle made by the phone, called preQ1. The preQ1 molecule changes the shape of the RNA when it binds to the riboswitch. This makes it possible for the RNAP to once again move along the DNA, allowing transcription to continue.
Although riboswitches were first identified in 2002, little is known about the specific functions they play in the transcription machinery. Expert on riboswitches Adrien Chauvier, a Walter Lab scientist, explains why this is the case.
He stated, “This is a David vs. Goliath situation.” RNAP is this monster, Goliath, and the riboswitch is David. Visualizing where and how preQ1 regulates transcriptional pausing is like finding a needle in a haystack due to the significant size difference.”
Prior research from the Walter lab uncovered that transcriptional stopping is turned on and off here and there as a component of the preQ1 particle restricting to the riboswitch. Moving forward, the Walter lab collaborated with Ohi, a specialist in cryo-EM, to visualize the situation.
“The strength of doing science at the University of Michigan can be seen in this work. According to Ohi, who is also a professor of cell and developmental biology at the University of Michigan Medical School, “three labs with different expertise were able to form a multidisciplinary collaboration that led to an important and novel discovery.” These discoveries could never have been conceivable without this collaboration, alongside the ventures the college has placed into reinforcing cryo-EM and RNA science at U-M lately.”
Single-molecule cryo-EM can decide the designs of enormous protein edifices by building 3D models from a huge number of 2D pictures of particles frozen in various directions, uncovering structures that contain sub-atomic subtleties that give useful bits of knowledge.
The primary data from single-molecule cryo-EM certified the Walter lab’s previous discoveries, yet in addition, it uncovered a particular change looking like the riboswitch never seen before. The riboswitch twists to communicate with the RNAP and allow transcription to continue when the preQ1 molecule binds.
Through a collaboration with Frank, then a professor of biophysics and chemistry at the University of Michigan and an expert in computational modeling of RNAs, these observations were further rationalized and validated. The U-M collaborative team now has a more precise understanding of this riboswitch’s regulation of transcriptional pausing thanks to detailed 3D models.
Walter stated, “Now that we understand the entire process of riboswitch regulation, we can use that knowledge to specifically target these critical parts of bacterial life, hopefully preventing the coming crisis of multidrug-resistant bacteria.”
Scientists all over the world are looking for ways to combat bacteria that can’t be killed by antibiotics to avoid a global health crisis.
A promising objective for better-than-ever anti-infection agents are riboswitches, little stretches of RNA that direct a cycle important for the development of proteins by the bacterial cell. Riboswitches are tracked down only in microorganisms and could be focused on with anti-infection agents, so creatures or people are unaffected. Researchers may be able to develop drugs that disrupt the cellular machinery that produces necessary proteins once they have a complete understanding of how riboswitches function.
Using a combination of biochemistry, structural biology, and computational modeling, researchers at the University of Michigan’s Life Sciences Institute and the Department of Chemistry have now revealed how a particular riboswitch regulates its own synthesis.
Transcription is the first step in making a protein from the genetic code. The genetic information in DNA is copied into a strand of RNA by the enzyme RNA polymerase (or RNAP), which moves along the DNA. RNAP will go through a number of “pauses” as it waits for the cell to give it more instructions during this process. Scientists have been trying for a long time to figure out how this pause and restart happens, but antibiotics could use this as a perfect target.
Using a structural biology technique known as single particle cryo-electron microscopy (cryo-EM), the team, which was led by chemistry professor Nils Walter and collaborated with the labs of LSI professor Melanie Ohi and former U-M scientist Aaron Frank, was able to see for the first time how this transcriptional regulation occurs. Nature Structural & Molecular Biology has published their findings.
The Walter lab took a gander at a specific riboswitch that ties a particle made by the phone, called preQ1. The preQ1 molecule changes the shape of the RNA when it binds to the riboswitch. This makes it possible for the RNAP to once again move along the DNA, allowing transcription to continue.
Although riboswitches were first identified in 2002, little is known about the specific functions they play in the transcription machinery. Expert on riboswitches Adrien Chauvier, a Walter Lab scientist, explains why this is the case.
He stated, “This is a David vs. Goliath situation.” RNAP is this monster, Goliath, and the riboswitch is David. Visualizing where and how preQ1 regulates transcriptional pausing is like finding a needle in a haystack due to the significant size difference.”
Prior research from the Walter lab uncovered that transcriptional stopping is turned on and off here and there as a component of the preQ1 particle restricting to the riboswitch. Moving forward, the Walter lab collaborated with Ohi, a specialist in cryo-EM, to visualize the situation.
“The strength of doing science at the University of Michigan can be seen in this work. According to Ohi, who is also a professor of cell and developmental biology at the University of Michigan Medical School, “three labs with different expertise were able to form a multidisciplinary collaboration that led to an important and novel discovery.” These discoveries could never have been conceivable without this collaboration, alongside the ventures the college has placed into reinforcing cryo-EM and RNA science at U-M lately.”
Single-molecule cryo-EM can decide the designs of enormous protein edifices by building 3D models from a huge number of 2D pictures of particles frozen in various directions, uncovering structures that contain sub-atomic subtleties that give useful bits of knowledge.
The primary data from single-molecule cryo-EM certified the Walter lab’s previous discoveries, yet in addition, it uncovered a particular change looking like the riboswitch never seen before. The riboswitch twists to communicate with the RNAP and allow transcription to continue when the preQ1 molecule binds.
Through a collaboration with Frank, then a professor of biophysics and chemistry at the University of Michigan and an expert in computational modeling of RNAs, these observations were further rationalized and validated. The U-M collaborative team now has a more precise understanding of this riboswitch’s regulation of transcriptional pausing thanks to detailed 3D models.
Walter stated, “Now that we understand the entire process of riboswitch regulation, we can use that knowledge to specifically target these critical parts of bacterial life, hopefully preventing the coming crisis of multidrug-resistant bacteria.”
More information: Chauvier, A. et al, Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand, Nature Structural & Molecular Biology (2023). DOI: 10.1038/s41594-023-01002-x www.nature.com/articles/s41594-023-01002-x