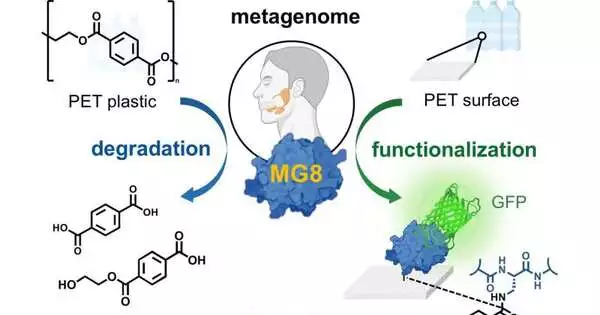
Human spit might contain a protein which can degrade the plastic polyethylene terephthalate (PET). Scientists tracked down the promising protein, a hydrolase, in a data set containing human metagenome tests. As they report in the journal Angewandte Chemie International Edition, this newfound hydrolase performs better compared to numerous other known bacterial PET hydrolases. It tends to be created utilizing biotechnological strategies and could be put to use in plastic recycling or for functionalizing plastics, the creators add.
Landfill locales and harbors are known to be especially encouraging destinations for finding microbes that have adapted to consuming or utilizing plastics. These microbes have advanced proteins known as PET hydrolases, which can separate PET into more modest atoms. Chayasith Uttamapinant from the Vidyasirimedhi Institute of Science and Technology (VISTEC) in Rayong, Thailand, and Worawan Bhanthumnavin from Chulalongkorn University, Bangkok, Thailand, and partners, have now found the main protein to decay PET from a fairly amazing source: the genome of microbial networks in human spit.
Analysts believe that because people consume large amounts of food packaged with PET, microorganisms in the spit, or the GI lot, have evolved to process microplastics.The group revealed the new hydrolase, which they named MG8, while looking through a public metagenome data set containing tests from seawater and human spit, and had the option to credit the logical wellspring of the protein to Gram-negative microbes that might live in human spit. These microbes are like strains seen as close to the “Pacific junk vortex,” which have additionally advanced to create PET hydrolases.
They originally required sufficient material to carry out their tests, so they changed a bacterium that can be refined in labs to create the protein. They handily recuperated a functioning type of the protein, ready to decay as PET, from a denatured structure that can be secluded in huge amounts. The analysts note that this shows extraordinary commitment to increase from now on.
Besides the possibilities for reusing versatility, the group also predicts one more utilization of MG8. They discovered that, in addition to being able to PET easily, it can also profoundly bind to it with a minor change. To accomplish this, they changed the protein grouping by supplanting one of the normally occurring amino acids (serine) at the dynamic site with an unnatural amino acid corrosive, DAP. The changed protein quickly stuck to the PET powder. This could be utilized as a vehicle for functionalizing PET surfaces, expanding the flexibility of PET in clinical gadgets, for instance, and improving the flexibility of reused PET.
In spite of the commitment of MG8 to plastic reusing and functionalization, the group recognizes that MG8, like other PET hydrolases, still has serious room for improvement. For now, buyer grade PET plastics with high crystallinity can’t be decayed utilizing this hydrolase. Hence, further exploration will be important to arrive at the stage where an entire plastic water jug can be broken up into a basic arrangement containing the protein.
More information: Bhumrapee Eiamthong et al, Discovery and Genetic Code Expansion of a Polyethylene Terephthalate (PET) Hydrolase from the Human Saliva Metagenome for the Degradation and Bio‐Functionalization of PET, Angewandte Chemie International Edition (2022). DOI: 10.1002/anie.202203061