Agents led by Neil Kelleher, Ph.D., teacher of medication in the Division of Hematology and Oncology and of organic chemistry and sub-atomic hereditary qualities, have fostered a computerized strategy for imaging and distinguishing proteoforms in ovarian disease tissue, as per results distributed in Nature Correspondences.
The strategy offers the best speed and exactness at present that anyone could hope to find for the high-goal, high-throughput imaging of proteoforms—every one of the changed variants of proteins in a tissue test—and has different likely applications in disease diagnostics, Kelleher said.
A few procedures are right now used to picture proteins in human tissue; however, not many are fit for imaging proteoforms. Those that can test proteoforms straightforwardly from tissue do so by ionizing them for mass spectrometry. Different methods keep researchers from understanding where the proteoforms exist inside a tissue and don’t recognize all current proteoforms in that frame of mind immediately.
“We have demonstrated in this paper that we can locate not just proteins but also their myriad proteoforms, the ultra-specific kind of measurement in my field of proteomics. This technique is really important for cancer tissue imaging and cancer diagnostics.”
Kelleher, who also directs the Proteomics Center of Excellence and the Chemistry of Life Processes Institute.
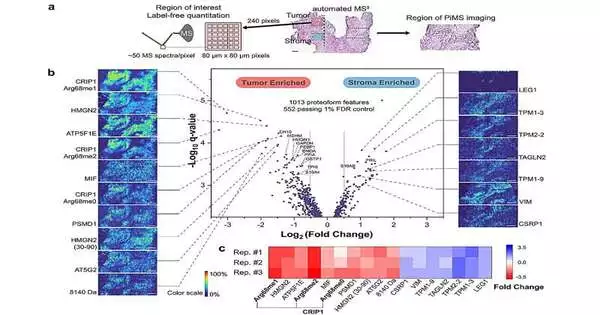
To all the more likely comprehend the spatial place of proteoforms inside a given tissue, Kelleher’s group created proteoform imaging mass spectrometry (PiMS), which was described point by point in a recent report distributed in Science Advances. The strategy works by examining proteoforms from the tissue with nanodroplets, “gauging” the removed proteoforms to distinguish those up to a specific size, and then utilizing this information to develop proteoform pictures of the filtered tissue.
In the ongoing review, Kelleher and his teammates based this procedure on AutoPiMS, which uses a computational motor to consequently distinguish and portray proteoforms inside a flimsy segment of malignant growth tissue.
AutoPiMS had the option to recognize and describe more than 300 proteoforms inside a human ovarian malignant growth tissue test and planned where explicit disease-related proteins existed inside the example at a speed of one moment for every proteoform. AutoPiMS was likewise ready to distinguish malignant growth tissues from non-carcinogenic tissues in a similar patient.
“This strategy is truly significant for malignant growth tissue imaging and disease diagnostics,” said Kelleher, who likewise coordinates the Proteomics Focus of Greatness and the Science of Life Cycles Foundation. “We have displayed in this paper that we can find proteins, however their heap proteoforms, the super unambiguous sort of estimation in my field of proteomics.”
The method will be made accessible to other Northwestern proteomics specialists, Kelleher said, and he trusts AutoPiMS will speed up disclosures in the field.
Pushing ahead, Kelleher and his partners will adjust the procedure for use in single-cell proteomics, he said.
“On the off chance that we can have data on single-cell protocorm’s, we would have the most exact data about proteins in space, time, and synthesis. That is a definitive innovation that would make protein investigation a lot more exact,” Kelleher said. “Accuracy medication requires accuracy proteomics. The more exact we can make it, the more we can propel drug advancement, lower aftereffects, and further develop diagnostics, which are all lined up with the new mission of the CLP Establishment at Northwestern.”
More information: John P. McGee et al, Automated imaging and identification of protocorm’s directly from ovarian cancer tissue, Nature Communications (2023). DOI: 10.1038/s41467-023-42208-3