A new report from Prof. Chong Kang’s group at the Chinese Academy of Sciences (CAS) Establishment of Organic Science has uncovered an original virus-tamed fix system for DNA harm in rice, comparing top modules to the improvement of the chilling resilience characteristic in rice with the codon rehashes at a solitary site.
The findings were published online in Science Advances.
Worldwide environmental change has prompted an unmistakable expansion in strange natural temperatures and outrageous climate events as of late. In this manner, it is imperative that harvests have the ability to persevere through outrageous temperatures to guarantee stable yields. Despite the fact that plants have developed muddled and fragile defensive instruments to endure chilling pressure, DNA harm actually happens, consequently bringing down plants’ safeguards. Moreover, the basic administrative instrument has been muddled to a great extent.
In this review, Prof. Chong’s gathering, in collaboration with Prof. Li Qizhai’s gathering from the Foundation of Arithmetic and Frameworks Study of CAS and Prof. Cheng Zhukuan’s gathering from the Establishment of Hereditary Qualities and Formative Science of CAS, utilized a methodology that joins populace hereditary qualities, genomics, and cell and developmental science to take advantage of the original module for chilling resilience.
The specialists combined information from broad affiliation studies (DM-GWAS) in light of multi-layered scaling. A progression of loci was distinguished by GWAS utilizing consolidated phenotypic information. One of these was qCTS11-1 on chromosome 11, which makes an unmistakable commitment to rice chilling resilience. Its significant quality, COLD11, was related to fine-scale planning. Loss-of-capability transformations of COLD11 caused decreased chilling resilience, as per the scientists.
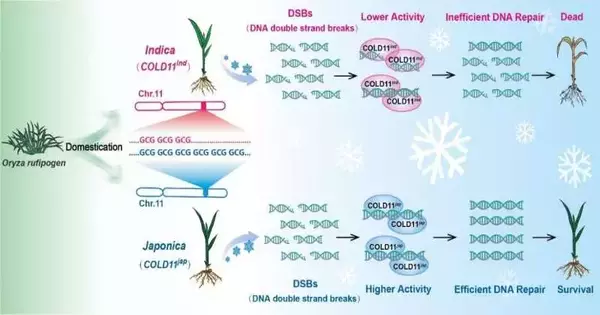
Various sorts of GCG codons that continue encoding alanine in the main exon of COLD11 were noticed for chilling-delicate indica assortments and chilling-open-minded japonica assortments. The GCG codon rehash numbers had an unmistakable, positive relationship with chilling resilience. Furthermore, genome development studies of agent rice germplasms revealed that different GCG arrangement rehashes revealed areas of strength for selection during the northern extension of rice planting.
Besides, COLD11 encodes a DNA-fixing protein that plays a fundamental role in the maintenance of DNA two-fold strand breaks. The GCG rehash numbers in its most memorable exon showed a positive connection with its biochemical movement. This is the primary report of a training selected DNA harm repair instrument and its comparison of top modules, including chilling pressure.
This study discovered that COLD11 is a significant quantitative characteristic loci quality for chilling resistance using DM-GWAS of Japonica and Indica—two rice subspecies with significant differences in chilling resilience.
The discovery of key codon rehashes in the main exon of COLD11, as confirmed by phylogenetic and geographic distribution analysis, paves the way for a fine guideline of rice chilling resistance with a solitary site. It is a possibly valuable sub-atomic module for further developing the chilling resistance characteristic in rice through the atomic structure.
More information: Zhitao Li et al, Natural variation of codon repeats in COLD11 endows rice with chilling resilience, Science Advances (2023). DOI: 10.1126/sciadv.abq5506. www.science.org/doi/10.1126/sciadv.abq5506