By combining AI with the laws of material science, analysts in the lab of Matthew Lew, academic partner of electrical and frameworks design at Washington College in St. Louis, have had the option of figuring out the direction and position of covering single particles in 5D from a solitary picture.
Their exploration was distributed on Sept. 26 in the journal Optics Express.
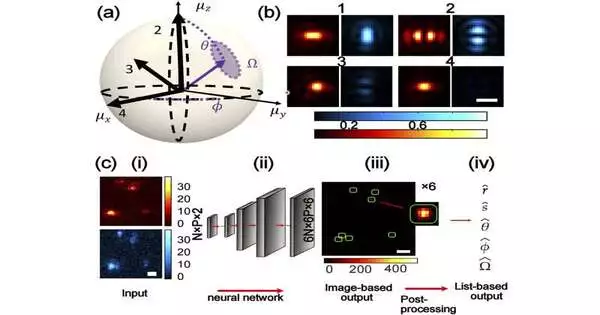
The five aspects being referred to aren’t new or secret spatial aspects. All things considered, a group headed by Tingting Wu, a Ph.D. understudy in the McKelvey School of Design’s imaging sciences program, had the option of planning a framework that could tell the direction of a particle in 3D space as well as its situation in 2D: five boundaries from a solitary, loud, pixelated picture.
Utilizing an AI calculation combined with post-handling permits the Lew lab to reveal the structure on the right from the loud, pixelated picture on the left. The picture on the right is variety coded with assessed 3D direction. Credit: Lew Lab
To wrest this extra, intricate data from an apparently basic spot of light, the group planned an AI calculation, yet added an additional step.
“A many individuals use man-made intelligence start to finish,” Wu said. “Just put in what you have and request that the brain network give you what you need.” She chose to break the issue into two moves to ease the burden on the calculation, making it stronger.
The sort of imaging done in the Lew lab—oof single atoms—wwill in general be “loud,” containing “bits” or changes that can cloud a picture. For most AI brain organizations, Lew said, “heartily managing that sort of clamor can be extremely muddled to learn.”
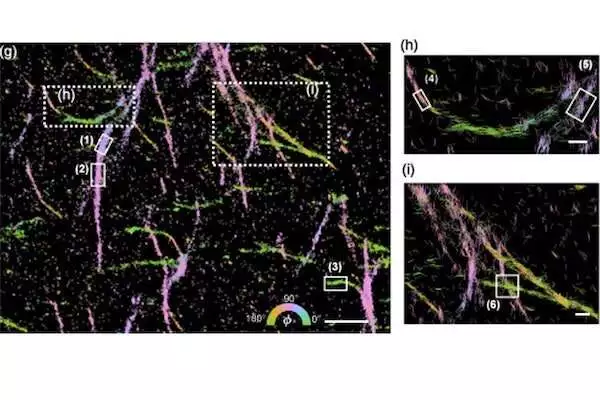
Amyloid proteins. Their lengths and bearings show the size of their in-plane directions and their directions, separately. Credit: Lew Lab
People, nonetheless, have previously figured out how signals from the atoms of interest and this clamor are joined together inside magnifying lens pictures. Rather than asking the calculation to re-get familiar with the laws of material science, the group added a second “post-handling” calculation—a clear calculation that applied these actual regulations to the outcomes from the main calculation.
“It resembles I’ve isolated two issues into two calculations,” Wu said.
Subsequent to handling a great many previews, the outcome, Wu said, is a “lovely picture” that utilizes tone, bend, and course to show how a huge number of particles are associated with one another.
At last, this framework will actually want to assist researchers with better grasping organic cycles at small scales, like the manner by which amyloid proteins gather to shape the tangled designs related to Alzheimer’s disease.
More information: Tingting Wu et al, Deep-SMOLM: deep learning resolves the 3D orientations and 2D positions of overlapping single molecules with optimal nanoscale resolution, Optics Express (2022). DOI: 10.1364/OE.470146
Journal information: Optics Express