By making it easier to administer pharmaceutical drugs precisely and promptly, the capability of rapidly identifying bacteria that are resistant to antibiotics has the potential to significantly contribute to the resolution of the global antibiotic crisis. The current diagnostic procedure for bacterial infections takes up to three days, making antibiotic treatment less effective.
Ann V. Nguyen and a Cornell University, New York, team developed a microfluidic system with a ladder-shaped design to generate a two-fold serial dilution of antibiotics that is suitable for national and international testing standards and was recently published in Communications Engineering. They were able to reduce the antibiotic susceptibility testing timeframe to approximately 4–5 hours thanks to the design and application processes. The findings of the study were in line with those of the kits that are currently on the market, making them a reliable and adaptable diagnostic tool for testing antibiotic susceptibility.
Antibiotic susceptibility testing and antibiotic resistance The worldwide effects of antibiotic resistance are attributed to the overuse of antibiotics in human and veterinary medicine, as well as flaws in animal handling. Overprescription of antibiotics, a lack of quick laboratory tests, and the distribution of prescription antibiotics that are only marginally effective, according to researchers, are to blame. Antibiotic resistance can be identified and characterized through the development and application of rapid diagnostic tests. The current work process for diagnosing antibacterial contamination typically requires up to 2–3 days, during which a patient goes through detachment, responsiveness, and recognizable proof.
In a clinical microbiology laboratory, the process can take up to two days for biochemists to identify the infectious agent based on the characteristics of the pathogen, which begins with the collection of samples. Specialists can segregate microorganisms to lead anti-microbial helplessness tests that take up another 16–20 hours, resulting in an absence of opportune procurement of the data on antibacterial opposition.
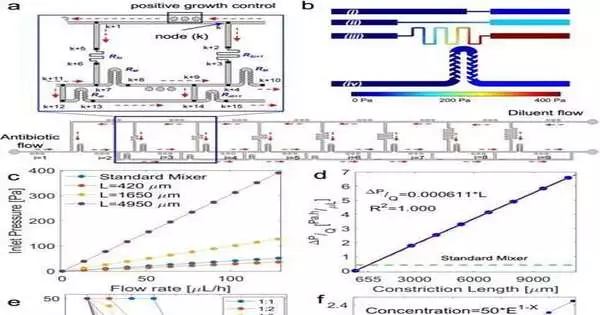
The pressure node network and computational fluid dynamics simulation a) The ladder flow distribution Before and after each resistor feature, as well as at the junctions, unique numbers are assigned to the pressure nodes in the inset. The flow’s direction is indicated by the red arrows. The counters for the resistors and nodes, respectively, are i and k. b) Pressure contours for constriction of (i) L = 420 m; (ii) L = 1650 m; (iii) L = 4950 m; and (iv) the standard mixer resistors for each channel’s flow rate of 130 L/h. c) The pressure at the features in A’s inlet d) The hydraulic resistance of the constriction, calculated as a function of length for a width of 40 millimeters. Reenactments for seven tightening influences are shown (L = 420 m, 1649 m, 4070 m, 4949 m, 7990 m, 10795 m). The constriction’s hydraulic resistance is the same as the mixer’s at L = 655 m. e) The concentration profile of the microchambers will be affected by the ratio of the drug and diluent flow rates at their respective inlets. Network simulations have produced these outcomes. f) In the concentration (%) = 50E1i formula, the dilution factor must be E = 2, where i is the chamber number. The least squares algorithm is used to solve for E by fitting the formula to the results in part E. Credit: Engineering of Communications (2023) DOI: 10.1038/s44172-023-00064-5
Microfluidic stages and the plan of a microfluidic stepping stool-like instrument
Bioengineers have subsequently evolved microfluidic stages as a technique to work on the responsiveness and speed of testing anti-microbial weaknesses. In this work, Nguyen and his team created a microfluidic system with an optimized ladder-like network for two-fold serial dilution distribution of culture media and antibiotics. They concentrated on the presentation of the stage to lead anti-infection testing and fostered a versatile, demonstrative instrument. The arrangement incorporated a microfluidic stage for anti-infection weakness testing, which consolidated nanoliter microchamber-based strategies with a stepping stool-like focus inclination generator.
They were able to quickly determine phenotypic antibacterial susceptibility thanks to the combination, which gave them a standard and adjustable antibiotic concentration profile. The platform was made by the researchers by bonding a PDMS layer to standard glass slides. This allowed them to test one antibiotic/bacterium combination per device. Throughout the experiments, each device was used as a bioreactor to incubate bacteria with antibiotics.
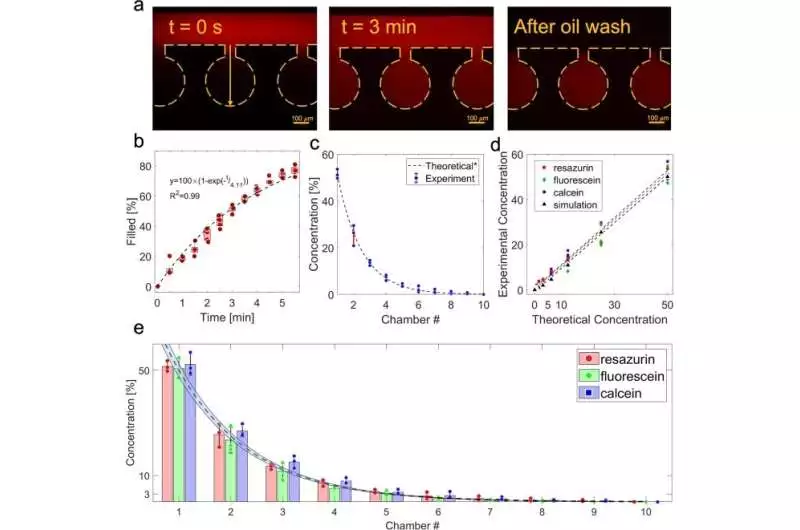
Portrayal of the fixation angle (a) pictures of the microchambers showing the course of particles diffusing into the microchambers from the fundamental channel at t = 0 s and t = 3 min, and after the primary channel is washed with oil. (b) The area under the curve of the concentration profile at each time point (n = 5) depicts the average concentration of resazurin that diffused into the microchambers over time. c) The concentration profile that emerged in ten of the ladder microfluidic system’s microchambers following a loading time of three minutes (n = 3). d) a linear correlation between the theoretical 2-fold dilution concentration profile and the computer-simulated concentration profiles of resazurin, fluorescein, and calcein created in the ladder microfluidic system. e) A comparison of the theoretical and specific loading times for the loaded concentration profiles of resazurin, fluorescein, and calcein. The run line with the blue zone shows the hypothetical fixation profile with a 5% blunder. The average standard deviation of all the data is displayed. Credit: Communications Engineering (2023). DOI: 10.1038/s44172-023-00064-5
The microfluidic instrument and its operating concept
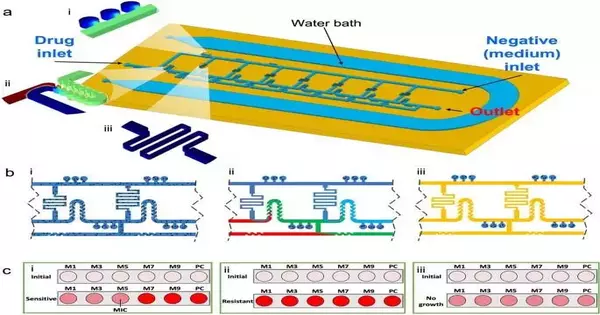
The device had three openings that formed a ladder-like structure: a drug inlet, a negative inlet, and an outlet. In order to dilute the antibiotics as they moved across the system from the drug inlet through a serpentine surface and mix them with bacteria at specific flow rates, the team added the antibiotics and culture media to the device from opposite directions. The bioengineers used computational fluid dynamics simulations to generate a wider range of dilutions in order to determine the precise flow rate. They then, at that point, investigated the standard of activity of the microfluidic framework with a previous convention.
The setup was constructed with a cation-adjusted Mueller-Hinton broth and the cell metabolism indicator PrestoBlue. To create a main channel network, they loaded the bacterial suspension into the device using a syringe, followed by an antibiotic solution. They looked at bacterial isolates from animal models and the performance of ladder microfluidic systems to figure out the minimum inhibitory concentration of antimicrobial substances on the platform. The instrument provided a system to notice various microbes, including Escherichia coli and Staphylococcus pseudointermedius. On the microfluidic cell-sorting platform, the bioengineers tested 206 bacterial samples in total.
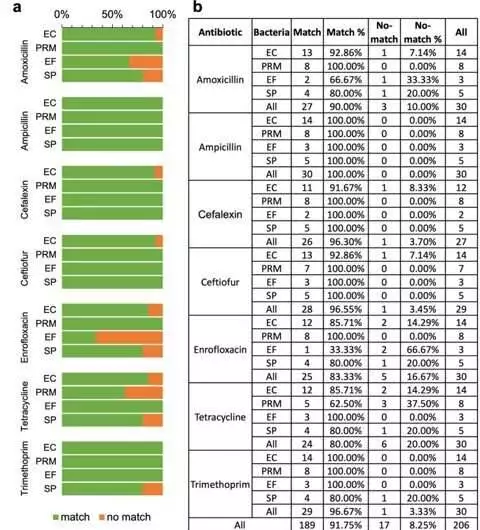
An examination of match versus unmatch between MICs got on-chip utilizing the stepping stool microfluidic framework and the highest quality level strategy directed at the veterinary symptomatic lab Escherichia coli (EC), Proteus mirabilis (PRM), Enterococcus faecalis (EF), and Staphylococcus pseudintermedius (SP) are among the targeted bacteria. b) A specific percentage of each antibiotic/bacterium combination that is matched versus unmatched The likelihood that each antibiotic-bacteria combination will yield an accurate MIC. Credit: Communications Engineering (2023). DOI: 10.1038/s44172-023-00064-5
Antibiotic susceptibility testing using spiked and clinical samples
The team looked into the possibility of using urine samples to test for antibiotic susceptibility without first isolating the bacteria. This was done with urine samples that were cultured and found to contain just one unknown bacteria. The group decided the limit with respect to negligible inhibitory focus by going through clinical examples in 4-5 hours.
A microfluidic system designed and optimized by Ann V. Nguyen and colleagues could be used to isolate bacteria from culture plates or directly from urine samples using phenotypic antibiotic susceptibility testing. In order to generate a two-fold concentration gradient, the instrument adhered to existing standardization procedures and utilized antibiotics and drug combinations that were clinically relevant. In order to generate a concentration gradient using microfluidics, the team used the instrument to load bacteria, antibiotics, and oil, and they also included a circuit logic system for the first time in the study. The method will be utilized by the researchers to expedite antibiotic susceptibility testing, thereby enhancing patient outcomes and streamlining clinical laboratory workflow in a short amount of time.
The team also wants to add automatic loading capacity and sample handling to the ladder microfluidic instrument so that it can do better real-time image analysis with better accuracy.
More information: Ann V. Nguyen et al, Ladder-shaped microfluidic system for rapid antibiotic susceptibility testing, Communications Engineering (2023). DOI: 10.1038/s44172-023-00064-5
Irith Wiegand et al, Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances, Nature Protocols (2008). DOI: 10.1038/nprot.2007.521