We live in a world made and run by RNA, the similarly significant kin of the hereditary atom DNA. Truth be told, developmental researchers estimate that RNA existed and self-duplicated even before the presence of DNA and the proteins encoded by it. Quick forward to current people: science has uncovered that under 3% of the human genome is interpreted into messenger RNA (mRNA) atoms that are converted into proteins. Conversely, 82% of it is deciphered into RNA particles with different capacities, a significant number of which actually stay baffling.
To comprehend what a singular RNA particle does, its 3D construction should be unraveled at the level of its constituent iotas and sub-atomic bonds. Scientists have regularly concentrated on DNA and protein atoms by transforming them into routinely pressed gems that can be analyzed with an X-beam bar (X-beam crystallography) or radio waves (atomic attractive reverberation). Notwithstanding, these procedures can’t be applied to RNA particles with almost similar adequacy on the grounds that their atomic structure and underlying adaptability keep them from effectively shaping precious stones.
At present, an exploration joint effort driven by Wyss Core Faculty members Peng Yin, Ph.D. at the Wyss Institute for Biologically Inspired Engineering at Harvard University, and Maofu Liao, Ph.D. at Harvard Medical School (HMS), has detailed a new way to deal with the underlying examination of RNA atoms on a very basic level. ROCK, as it is known, employs an RNA nanotechnological strategy that allows it to collect numerous indistinguishable RNA particles into an extremely coordinated structure, which completely decreases the adaptability of individual RNA atoms and duplicates their sub-atomic weight.to notable model RNAs with various sizes and capacities as benchmarks, and the group showed that their strategy empowers the primary examination of the contained RNA subunits with a method known as cryo-electron microscopy (cryo-EM). Their development is accounted for in Nature Methods.
“Cryo-EM has significant advantages over traditional methods for viewing high-resolution details of biological molecules such as proteins, DNAs, and RNAs, but the small size and moving tendency of most RNAs prevents successful RNA structure determination. Our novel method of assembling RNA multimers addresses both of these issues simultaneously by increasing the size of RNA while decreasing its movement. Our approach has paved the way for cryo-EM structure determination of many RNAs. The team named their method “RNA oligomerization-enabled cryo-EM via kissing loops” because it combined RNA nanotechnology and cryo-EM approaches (ROCK)”.
Liao, who is also an Associate Professor of Cell Biology at HMS,
Yin, who led the review with Liao, stated that Rock is breaking the ongoing limits of RNA underlying examinations and enables 3D designs of RNA particles to be opened that are difficult or difficult to access with existing techniques and at close nuclear target.”We anticipate that this advance should strengthen numerous areas of key exploration and medication improvement, including the blossoming field of RNA therapeutics.” Yin is additionally the head of the Wyss Institute’s Molecular Robotics Initiative and Professor in the Department of Systems Biology at HMS.
Dealing with RNA
Yin’s group at the Wyss Institute has spearheaded different methodologies that empower DNA and RNA atoms to self-gather into enormous designs in light of various standards and prerequisites, including DNA blocks and DNA origami. They hypothesized that such techniques could also be used to gather naturally occurring RNA atoms into highly requested roundabout structures in which their ability to flex and move is severely limited by explicitly connecting them together.Numerous RNAs fold in complex yet unsurprising ways, with little sections base-matching with one another. “Profoundly” and “stem-circles” protrude out into the fringe.
“In our methodology, we introduce ‘kissing circles’ that connect different fringe stem-circles having a place with two duplicates of an indistinguishable RNA in a manner that permits a generally settled ring to be framed, containing numerous duplicates of the RNA of interest,” said Di Liu, Ph.D., one of two first-creators and a postdoctoral fellow in Yin’s group. “We hypothesized that these higher-request rings could be examined with a high goal by cryo-EM, which had been applied to RNA particles with first achievement.”
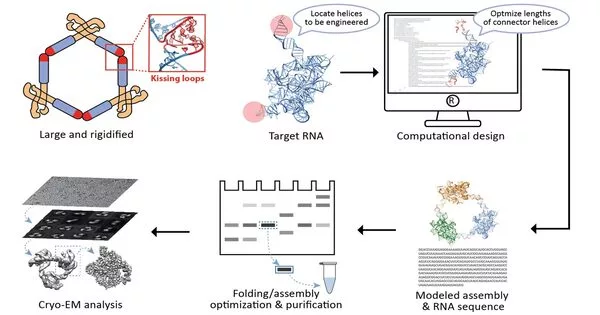
Imagining settled RNA
With cryo-EM, many single particles are streak frozen at cryogenic temperatures to forestall any further developments and afterward envisioned with the assistance of an electron magnifying instrument and the assistance of computational calculations that think about the different parts of a molecule’s 2D surface projections and reproduce its 3D design. Peng and Liu collaborated with Liao and his previous alumni understudy, François Thélot, Ph.D., the other co-first creator of the review. Liao, with his team, has made significant commitments to the quickly progressing cryo-EM field and the exploratory and computational examination of single particles shaped by unambiguous proteins.
“Cryo-EM has a significant advantage over conventional techniques for observing high-goal subtleties of organic particles, including proteins, DNAs, and RNAs; however, the small size and moving inclination of most RNAs preclude effective assurance of RNA structures.” clever strategy for gathering RNA multimers takes care of these two issues simultaneously, by expanding the size of RNA and decreasing its development,” said Liao, who is additionally an Associate Professor of Cell Biology at HMS. “Our methodology has paved the way for quick design assurance of numerous RNAs by cryo-EM.” The incorporation of RNA nanotechnology and cryo-EM approaches drove the group to name their technique “RNA oligomerization-empowered cryo-EM by means of introducing kissing circles” (ROCK).
To give verification of-standard to ROCK, the group zeroed in on an enormous intron RNA from Tetrahymena, a solitary-celled organic entity, and a little intron RNA from Azoarcus, a nitrogen-fixing bacterium, as well as the purported FMN riboswitch. Intron RNAs are non-coding RNA successions dissipated all through the groupings of newly deciphered RNAs and must be “joined” out for the adult RNA to be created. The FMN riboswitch is found in bacterial RNAs associated with the biosynthesis of flavin metabolites derived from vitamin B2. After restricting one of them, flavin mononucleotide (FMN), it switches its 3D adaptation and stifles the union of its mom RNA.
“The get-together of the Tetrahymena bunch I intron into a ring-like construction made the examples more homogenous and empowered the utilization of computational devices utilizing the balance of the collected design.” “While our dataset is generally small in size, ROCK’s natural advantages enabled us to determine the construction at a phenomenal goal,” Thélot explained.The RNA’s center is settled at 2.85 [one ngström is one ten-billionth of a meter and the favored measurement utilized by underlying biologists], uncovering definite elements of the nucleotide bases and sugar spine. I don’t figure we might have arrived without ROCK — or possibly not without significantly more assets. “
Cryo-EM can also catch atoms in various states in the event that they, for instance, change their 3D compliance as a feature of their capacity. Applying ROCK to the Azoarcus intron RNA and the FMN riboswitch, the group figured out how to recognize the various adaptations that the Azoarcus intron changes through during its self-joining process and to uncover the relative conformational unbending nature of the ligand-restricting site of the FMN riboswitch.
“This concentrate by Peng Yin and his associates carefully demonstrates the way that RNA nanotechnology can fill in as a gas pedal to progress different disciplines.” “Having the option to imagine and comprehend the designs of many normally occurring RNA atoms could hugely affect how we might interpret numerous natural and obsessive cycles across various cell types, tissues, and life forms, and, surprisingly, empower new medication improvement as it draws near,” said Wyss Founding Director Donald Ingber, M.D., Ph.D.