An examination group from Kiel College depicts an obscure guard system in microbes that specifically avoids unfamiliar and possibly unsafe hereditary data.
Since the COVID pandemic, the especially fast developmental versatility of microorganisms like microbes or infections has been brought into the public spotlight. For instance, when infections foster the capacity to taint new host creatures or microbes with anti-toxin opposition, the take-up of new hereditary data from different microorganisms permits them to communicate developmentally favorable qualities rapidly.
Microbes, for instance, take up unfamiliar DNA through a cycle called flat quality exchange, which is a lot quicker than the upward legacy from one generation to another.
Nonetheless, every living creature likewise faces gambles by taking up unfamiliar hereditary data, as it might actually be risky if, for instance, significant qualities are harmed by mixing into its own chromosome, bringing about significant burdens for the organic entity overall. Hence, microbes have developed various systems shielding them from retaining unsafe DNA. A large number of the atomic cycles included have recently been discovered, prompting the coinage of the phrase “bacterial safe framework.”
“Bacteria use plasmids to obtain new, non-critical genetic information. As a result, it is evident that a defense mechanism must be selective and not destroy all plasmids.”
Professor Marc Bramkamp
Presently, a group from the Microbial Natural Chemistry and Cell Science Gathering at the Foundation of General Microbial Science at Kiel College has explained the capability of another guard system that can recognize and, if vital, separate specific free and portable DNA structures called plasmids in bacterial cells — while recognizing helpful and unsafe hereditary data.
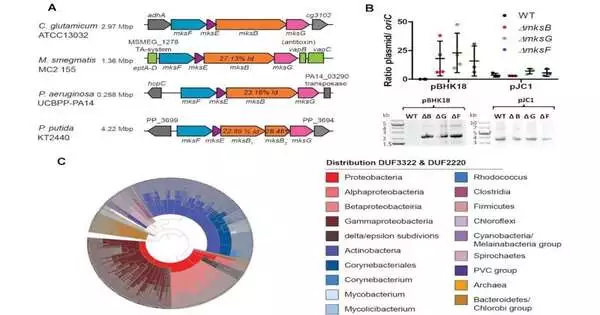
Utilizing the bacterium Corynebacterium glutamicum, for instance, the scientists showed that the alleged Mks protein framework has an extra component that can tie to plasmid DNA and cut it apart. The Kiel researchers, led by Teacher Marc Bramkamp, distributed their new outcomes in Nucleic Acids Exploration.
Proteins for DNA association can likewise guard against plasmids.
Plasmids are little, normally ring-molded, twofold-abandoned DNA atoms that can repeat freely on the chromosome in their host cell. They assume a significant role in the nature and development of microbes, as they are a significant vehicle of parallel quality exchange, empowering the fast exchange of hereditary data and hence the outflow of choice benefits. On a basic level, all microbes can trade plasmids with one another, even across animal types.
This happens straightforwardly from one bacterium to another through an exchange system known as formation. Both favorable and disadvantageous plasmids use such scaffolds between bacterial cells to change, starting with one bacterium and moving on to the next.
“How the bacterial creature manages unfamiliar DNA from recently moved plasmids has been little explored up to this point,” Manuela Weiß, a Ph.D. understudy in Bramkamp’s examination bunch, calls attention to. “In past exploration, we have examined frameworks that are by and large engaged with the association of DNA in bacterial cells and, in addition to other things, guarantee the bundling of hereditary data into the packed type of chromosomes,” Weiß proceeds.
In this unique situation, the examination group got early signs that C. glutamicum has two such frameworks, one of which isn’t engaged with the association of the chromosome yet can forestall the increase of specific plasmids, albeit the system liable for this was already obscure.
Presently, the Kiel analysts, along with specialists led by Dr. Anne Marie Wehenkel from the Institut Pasteur in Paris, have found the DNA scissors of the Mks framework in a primary report. “We had the option to demonstrate tentatively that this new subunit of the MKS framework shapes a particular protein, a supposed nuclease, which can cut DNA. This component has the mission of debasing plasmids to get unsafe DNA far from the bacterial cell, while different parts of the MKS framework are significant for the acknowledgment of plasmid DNA,” Weiß says.
Recognizing helpful and unsafe plasmids
The scientists then circled back to the perception that the MKS framework clearly just debases specific plasmids and that it should hence be connected to a choice system. A significant benefit here is that Bramkamp’s exploration team is working with the bacterium C. glutamicum, a creature that normally has this framework. Its capabilities can therefore be concentrated in vivo without its organic properties being changed by moving it into a model framework.
“Microbes utilize specific plasmids as a wellspring of new, not quickly crucial, hereditary data.” “It is hence clear that a guard system should be specific and not obliterate all plasmids,” says Bramkamp.
“We had the option to demonstrate that in C. glutamicum there is for sure a coordinated choice as per useful and negative hereditary data. At the point when we falsely turned off the MKS framework and hence all plasmids stayed in the bacterial cells, adverse impacts on the cell, perhaps set off by DNA stress, were clear. Nevertheless, these didn’t happen when the guard system was dynamic,” Bramkamp proceeds.
With the flow work, the Kiel analysts are introducing significant new discoveries about the bacterial safe framework generally, which grow the comprehension of plasmids as arbiters of useful as well as unsafe hereditary data. In the future, they need to explore which atomic systems permit bacterial cells to separate “great” and “awful” portable DNA.
The new outcomes are not just significant for the general comprehension of the association and generation of bacterial life. The undeniably exact examination of the bacterial safe framework could likewise assist with bettering address applied difficulties — and, for instance, more readily show and anticipate the advancement of anti-toxin opposition in specific bacterial populations later on.
More information: Manuela Weiß et al, The MksG nuclease is the executing part of the bacterial plasmid defense system MksBEFG, Nucleic Acids Research (2023). DOI: 10.1093/nar/gkad130