Researchers from the USC Michelson Center for Convergent Bioscience have created a ground-breaking method that offers a new method for rapidly gathering and organizing extremely detailed information about organic tissues.
Future applications could include the quick processing of tissue biopsies for cancer treatment or the detection of bacteria in food processing facilities.
While detectable, the intrinsic fields that tissues emit are very weak and difficult to distinguish. The method makes use of a sophisticated mathematical algorithm to enhance the signal quality before separating it, as described in two papers that were published in Nature Methods and Cell Reports Methods.
According to Francesco Cutrale, co-principal investigator and research assistant professor at the USC Viterbi School of Engineering, the new method is comparable to how a streaming service presents various levels of compression to guarantee their video is consistent regardless of a user’s internet connection.
“We’re doing something similar: we’re taking really massive, extremely complex data and compressing it. We may then examine very big data sets, which are linked by similarity into a massive histogram, and analyze them in record time and with extremely high sensitivity.”
Francesco Cutrale, co-principal investigator and research assistant professor at the USC Viterbi School of Engineering.
The streamer will send the video with various levels of compression, which are then recomposed for your device according to how quickly your connection can handle them, he explained. “We’re doing something similar: moving extremely large, extremely complex data into a space where it is compressed. Then, we can examine very large data sets that are grouped together into a massive histogram based on similarity, and we can perform extremely sensitive and quick analyses on the data.
A 20-frame-per-second SHy-Cam image of a beating Zebrafish embryo’s heart. The University of Southern California is to be credited.
A peek at the complexities of cells and biological tissue
The algorithm, which was described in more detail in Nature Methods earlier this year, is a continuation of the recent improvement of fluorescence-based high-content imaging techniques. Fluorescence has made it possible to identify and detect specific molecules due to its high specificity, contrast, and adaptability. These more recent methods, however, are ineffective for imaging in vivo, or live, samples due to their low sensitivity and potential for specimen damage.
The research team demonstrated in the paper how the technique, known as hybrid unmixing, could be used to accurately and quickly analyze live organic tissue. In order to analyze various components within a specimen marked by chemical substances known as fluorophores, a technique called linear unmixing is used.
They then employ hyperspectral phasors, which employ the entire color spectrum rather than just red, blue, and green, to visualize these components. Hybrid unmixing accomplishes this by enabling simultaneous imaging of bright and dimly labeled components within organic tissue, even in low-light conditions.
The method will give more precise insights into the complexity of biological systems by enabling concurrent analysis of the cellular behaviors and cellular metabolism of these labeled components.
According to Cutrale, complex biological systems are being studied more in the research community.
“Despite the fact that researchers frequently only look at two or three labels at once, there are actually numerous factors interacting within cells. It’s difficult to distinguish between these signals because they frequently have a similar appearance. Our paper successfully separates and identifies up to 14 different signals. This discovery will give researchers a more thorough understanding of how biological and cellular systems function.
According to Cutrale, the algorithm lays the groundwork for a wide range of applications in the industry.
“We work in the life sciences, but it’s easy to imagine numerous applications to evaluate the quality of fruits, the presence of pesticides, or how to optimize production in many other fields,” he said.
SHy-Cam provides an affordable, high-quality imaging tool.
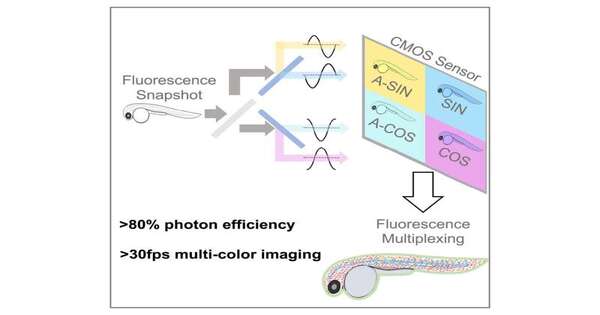
The research team developed hardware called SHy-Cam, short for Single-shot Hyperspectral Phasor Camera, that is described in a subsequent paper that was just released in Cell Reports Methods. SHy-Cam was created with the goal of capturing this kind of data. Fluorescence-based tissue imaging methods frequently use a variety of color channels to account for label overlap. The speed of imaging is slowed down by this method, and if the samples are exposed to too much light, damage may result.
The researchers were able to quickly and effectively gather spectral data with the SHy-Cam, a camera that can be constructed from readily available optical parts, using the new algorithm. The new apparatus described in the paper has a photon efficiency of over 80% and can acquire 30 data sets per second. According to the researchers, this makes it an effective tool for multi-color in vivo imaging.
Cutrale stated, “How do you produce a two-dimensional picture with a 2D sensor? You take a picture. ” Our problem is how to acquire a 3D data set with a 2D sensor. Red, blue, and green are the typical colors that a color sensor picks up, or it can use its grayscale sensors to collect all the information.
“In our case, we need to request 42 channels of information, which is unusual and ineffective. In this paper, we present a novel method for obtaining a spectral encoded version from a single image.”.
They employ light, according to Cutrale, to accomplish this. Prior to compressing the data onto the sensor, the team used light to transform the information and perform the calculations. The team demonstrated how this method can be used to receive the entire spectrum as well as the image’s dimensions.
In a single image created using a standard camera, “we have captured the X- and Y-axes of the image—its height and width—as well as the spectral information on the wavelength axis,” he claimed. “That strategy is quite effective. We were able to achieve hardware efficiencies that are, in some cases, up to eight times faster than existing instrumentation. In this compressed manner, eight times more light enters the camera sensor.
More information: Francesco Cutrale, a Single-shot Hyperspectral Phasor Camera for fast, multi-color fluorescence microscopy, Cell Reports Methods (2023). DOI: 10.1016/j.crmeth.2023.100441. www.cell.com/cell-reports-meth … 2667-2375(23)00056-5
Hsiao Ju Chiang et al, HyU: Hybrid Unmixing for longitudinal in vivo imaging of low signal-to-noise fluorescence, Nature Methods (2023). DOI: 10.1038/s41592-022-01751-5