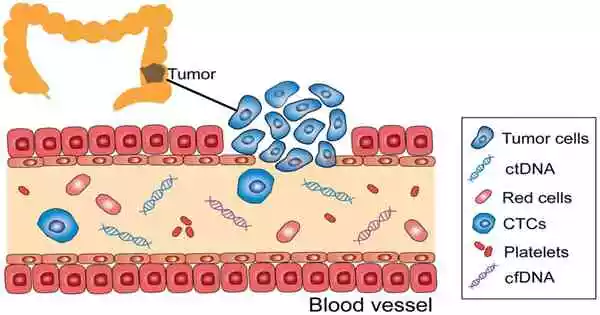
Pathogen detection, genetic disease identification, and early cancer diagnosis are the most common applications for nucleic acid analysis. For example, quantitative analysis of circulating tumor DNA (ctDNA), a free DNA fragment produced by malignant cells that contains tumor-specific sequence modifications, can assist in obtaining a wealth of information about tumors, such as gene point mutations and genome integrity. As a result, ctDNA is regarded as a customized tumor marker and plays an important role in cancer diagnosis and malignancy assessment.
Miao Peng’s group at the Chinese Academy of Sciences’ Suzhou Institute of Biomedical Engineering and Technology (SIBET) recently created an electrochemical DNA nanomachine based on a looped bipedal DNA walking reaction for very sensitive nucleic acid measurement. The pertinent findings were reported in ACS Central Science.
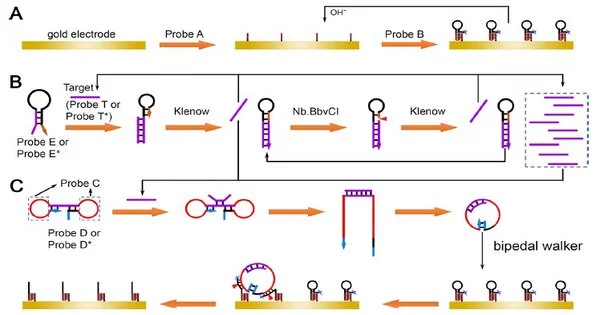
Miao and his colleagues used sequence design to create a pH-controllable intermolecular triple helix DNA nanostructure between DNA probes A and B, and then used that structure to create a renewable modified electrode interface.
To magnify the information in a target sequence, they devised a simple yet effective strand displacement amplification approach. “The reaction rate was effectively improved by integrating the primer and template into a single hairpin-structured DNA probe,” Miao explained.
In the presence of a target sequence, a high number of single-stranded DNA products could be created.
“Through the integration of primer and template into a single hairpin structured DNA probe, the reaction rate was effectively improved,”
Miao Peng’s
They also devised a revolutionary looped bipedal DNA walking method. The two loops of the dumbbell-shaped DNA probe had DNAzyme sequences that could not initially react with the track strands at the electrode contact (inactivated state).
When activated by the single-stranded DNA created by the preceding strand displacement amplification, it formed a looped structured DNA probe from the dumbbell probe. The bipedal walkers were engaged in further engagement with the track strands at the electrode surface, causing electrochemical reaction alterations.
According to their findings, under ideal experimental conditions, 2.2 aM (approximately 1.3 copy/L) sensitivity could be attained using the devised approach. “This approach has good selectivity,” Miao remarked.
Clinical serum samples and throat swab samples were analyzed and verified further.
According to Miao, the corresponding patients can be efficiently distinguished from the healthy control groups by analyzing aberrant electrochemical signals.
In addition, the concept provides a new rapid and sensitive method for detecting DNA markers of acute infectious illnesses.