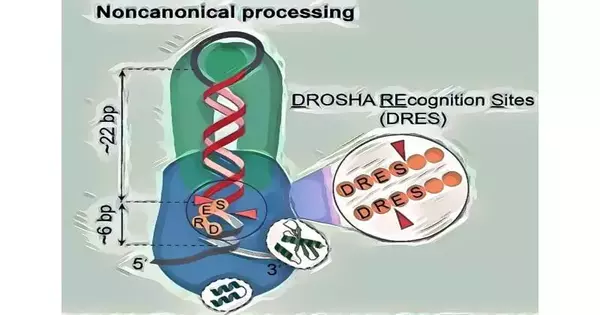
The Hong Kong University of Science and Technology (HKUST) research team, led by Assistant Professor of the Division of Life Science Prof. Tuan Anh Nguyen, utilized a number of sophisticated methods, including miRNA sequencing, pri-miRNA structure analysis, and high-throughput pri-miRNA cleavage assays for approximately 260,000 pri-miRNA sequences, in order to thoroughly discover and demonstrate the newly discovered noncanonical cleavage mechanism.
The noncanonical mechanism, in contrast to the canonical mechanism, does not rely on the essential protein and RNA elements that the canonical mechanism requires. The concentrate additionally uncovered previously unnoticed DROSHA acknowledgment locales (DRES), which are basic for noncanonical cleavage but can likewise work in the sanctioned cleavage component.
Moreover, the review features the developmental part of this noncanonical cleavage component, uncovering that it is moderated across a few animal categories. This suggests that the evolution of miRNA biogenesis and regulation is significantly influenced by noncanonical mechanisms.
Small RNA molecules known as microRNAs (miRNAs) play a crucial role in regulating gene activity. They control cell growth, development, and immunity, among other biological processes. Lately, researchers have been exploring miRNAs broadly to better comprehend their capabilities and the components involved in their creation.
A noncanonical cleavage mechanism for the Microprocessor (MP, DROSHA-DGCR8 complex) complex, which is responsible for producing miRNAs in humans and other animals by processing primary miRNA transcripts (pri-miRNAs), has recently been discovered by researchers at the Hong Kong University of Science and Technology (HKUST).
This ground-breaking discovery sheds light on a long-standing molecular biology mystery and may have significant repercussions for our comprehension of gene regulation, cellular processes, and the development of animal miRNA biogenesis pathways.
The microchip complex in creatures was found in 2004, and from that point forward, its sub-atomic systems for delivering miRNAs have been broadly concentrated on by many exploration gatherings. The canonical pri-miRNA processing mechanism is the model of this enzyme’s molecular mechanism that was created by these studies together.
Nonetheless, this component can make sense of how the compound cuts numerous miRNAs in creatures. This mechanism fails to explain how a significant portion of pri-miRNAs are processed because animal pri-miRNAs have very different structures and sequences.
A two-decade-old mystery in molecular biology regarding the cleavage of numerous pri-miRNAs in animals is solved by the noncanonical pri-miRNA processing mechanism, which was discovered and published in Molecular Cell. It also adds to our knowledge of the canonical mechanism. To put it more succinctly, this discovery reveals a novel method by which our cells produce miRNAs. This could have an impact on our comprehension of gene regulation, cellular processes, and the development of animal miRNA biogenesis pathways.
Among the study’s most important findings are:
- The revelation and far-reaching portrayal of another way cells produce miRNAs in creatures settles a well-established secret in sub-atomic science.
- The discovery of DRES that controls MP’s cleavage efficiency and accuracy opens up a new area of inquiry into the possibility of RNA recognition sites in other enzymes that are similar.
- There is evidence that the noncanonical miRNA production mechanism, which plays a significant role in miRNA biogenesis in worms like C. elegans and C. briggsae, is shared by a number of animal species.
- An explanation for how MP cleaves numerous short stem miRNAs in animals suggests that the MP complex has broader cellular functions.
The disclosure of the noncanonical cleavage system of the MP complex in miRNA biogenesis has extensive ramifications for future examinations in sub-atomic science. The discovery of this mechanism broadens our comprehension of the regulatory landscape of miRNA biogenesis in animals and opens up new research directions.
One of the main ramifications is the possibility of finding a more prominent assortment of substrates for the MP complex in creatures. Previously, the processing of some pri-miRNAs could not be explained by the canonical mechanism. The noncanonical mechanism now enables researchers to reexamine previously overlooked or unexplained RNA substrates. This could prompt the recognizable proof of novel pri-miRNAs and other RNA substrates that are explicitly handled by the noncanonical component.
The possibility of uncovering novel animal functions for the MP complex is another implication. As the noncanonical component can handle short stem pri-miRNAs, it recommends a more extensive cell job for the MP complex. As a result, it’s possible that new roles in gene regulation and cellular processes like development, differentiation, and immunity will emerge.
Lastly, this study highlights the evolutionary significance of this mechanism by demonstrating that the noncanonical mechanism is conserved across a variety of animal species, particularly in worms like C. elegans and C. briggsae. The evolution and diversification of animal miRNA biogenesis pathways could be illuminated by further research into the evolutionary aspects of both canonical and noncanonical mechanisms.
In conclusion, our comprehension of the molecular mechanisms underlying miRNA production has been enhanced, and new research avenues have emerged as a result of the discovery of the noncanonical mechanism in miRNA biogenesis. This opens the door to additional discoveries in molecular biology, which will improve our comprehension of cellular processes, gene regulation, and the development of animal miRNA biogenesis pathways.
More information: Thuy Linh Nguyen et al, Noncanonical processing by animal Microprocessor, Molecular Cell (2023). DOI: 10.1016/j.molcel.2023.05.004