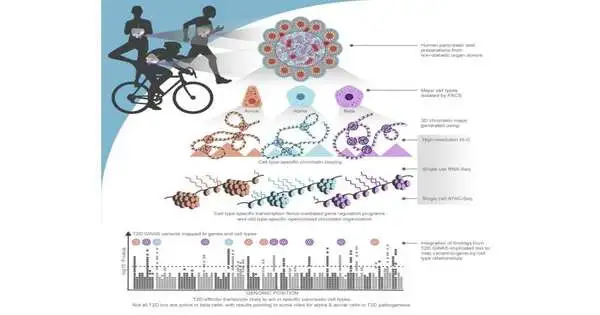
Scientists from Children’s Hospital of Philadelphia (CHOP) have utilized advanced three-layered planning procedures at a minute level to distinguish a large number of hereditary variations and relate objective quality pairings in the pancreas that are embroiled in type 2 diabetes. Notwithstanding these revelations, the subsequent datasets will act as a secret weapon for specialists all around the world to dive further into the hereditary starting points of type 2 diabetes and further investigate the roles of various sorts of cells in the improvement of the illness.
The discoveries were published today in the journal Cell Metabolism.
Type 2 diabetes cases are on the ascent and being analyzed in patients earlier in life than what has been generally noticed. Nonetheless, while many cases can be credited to an ascent in stoutness and a stationary way of life, expanding proof recommends serious areas of strength for a hereditary gamble factor that plays in this somewhat normal illness. Earlier broad affiliation studies (GWAS) have recognized many hereditary variations related to an expanded risk of creating type 2 diabetes.
One of the difficulties in exploring hereditary variations is to imagine chromosomes working while stuffed thickly inside minute cells. Assuming the DNA from one cell was loosened up start to finish, it would be in excess of six feet in length. Protein edifices called chromatin act to overlap chromosomes and reduce them to every other phone. Hence, qualities and variations that seem far off and irrelevant in a one-layered guide of the genome grouping can be firmly related in 3D in a cell, assisting with making sense of the basis of how hereditary sicknesses are caused.
“In this study, we used three-dimensional mapping techniques to chromosomes of highly significant cell types, which allowed us to find a set of genes that had never previously been involved in type 2 diabetes.”
Andrew D. Wells, Ph.D., an Associate Professor of Pathology and Laboratory Medicine
In the event that you don’t have a precise guide of how qualities are collapsed together in a cell, you’re basically speculating the job that these hereditary variations play,” expressed one of the review’s senior creators, Andrew D. Wells, Ph.D., an Associate Professor of Pathology and Laboratory Medicine and Director of the Spatial and Functional Genomics Collaborative at CHOP. “In this review, we applied three-layered planning procedures to chromosomes of exceptionally pertinent cell types, which permitted us to distinguish a progression of qualities that have never been embroiled before in type 2 diabetes.”
In this review, scientists made three-layered epigenomic profiles of sanitized acinar, alpha, and beta cells in the human pancreas, utilizing different strategies that analyzed every one of them at the single-cell level. While contrasting these profiles, the specialists found contrasts in how chromatin was compartmentalized, how the chromatin was circled, and the way that qualities were being managed and deciphered. Thus, the review group recognized a progression of causal variations and target quality matches at 194 different sorts of type 2 diabetes signals utilizing the three-layered chromatin maps. Moreover, the review uncovered that alpha and acinar cells probably play a larger part in the improvement of type 2 diabetes than recently suspected.
One of the limits of the paper is that these recently embroiled quality variation pairings were not completely approved in that frame of mind prior to examinations. The objective of this paper was to give basic data so various labs could additionally investigate and approve these revelations.
“Earlier examinations seem to help large numbers of the discoveries we made during the time spent this work, and with extra approval and imaging devices, more forceful drivers of type 2 diabetes pathogenesis at a hereditary level can be detached,” expressed one of the review’s senior creators, Struan F.A. Award, Ph.D., the other Director of the Center for Spatial and Functional Genomicsand the Daniel B. Burke Endowed Chair for Diabetes Research at CHOP. “We accept the discoveries of this study will act as a vital asset for the diabetes research local area.”
The other senior creator was Klaus Kaestner, from the Department of Genetics at the University of Pennsylvania.
More information: Chun Su et al, 3D chromatin maps of the human pancreas reveal lineage-specific regulatory architecture of T2D risk, Cell Metabolism (2022). DOI: 10.1016/j.cmet.2022.08.014
Journal information: Cell Metabolism