A new evolutionary model that suggests a billion-year evolutionary arms race between two groups within the Bamfordvirae virus kingdom and their hosts has been proposed by researchers.
The editors of their study, which was published today as a peer-reviewed preprint in eLife, say that it provides “convincing” analyses that improve our comprehension of the deep evolutionary history of viruses, the interaction between viruses and the first eukaryotes (organisms with cells that include a nucleus), and the diversity of viral lineages.
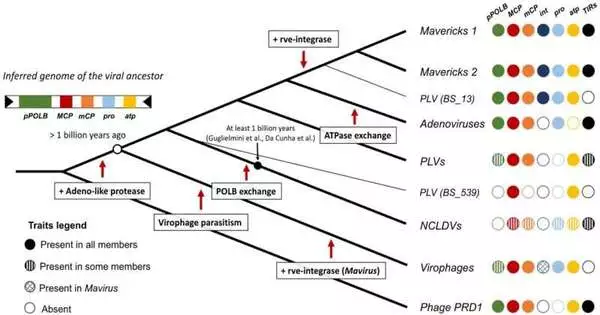
One of the most diverse groups of viruses that infect living things is the family Bamfordvirae. They incorporate nucleocytoplasmic huge DNA infections (NCLDVs; the biggest infections described to date), virophages (viral parasites of other infections), adenoviruses (normal infections that cause cold and influenza-like side effects), and protesters and Polinton-like infections (both infections with versatile hereditary components that colonize the genomes of their hosts).
“Despite these hypothesized possibilities, the development of viruses in the Bamfordvirae kingdom continues to raise significant unanswered questions. We intended to test the predictions of both the nuclear-escape and virophage-first theories, and investigate other hypotheses on the genesis of certain lineages, in order to better understand their history.”
José Gabriel Niño Barreat, Postdoctoral Research Assistant at the University of Oxford, UK.
There are two fundamental speculations for the starting points of these infections: the “virophage-first” and “nuclear-escape” hypotheses. According to the nuclear-escape hypothesis, a Maverick-like ancestor escaped from the nucleus of the host cell and gave rise to adenoviruses and NCLDVs after originating with hosts (endogenous). The virophage-first hypothesis, on the other hand, suggests that NCLDVs evolved alongside earlier virophages. After that, adenoviruses escaped from the host nucleus at a later stage, and mavericks evolved from virophages that became endogenous.
“In spite of these proposed situations, the broadening of infections in the Bamfordvirae realm remains a significant open inquiry in infection development. “We wanted to test the predictions made by both the nuclear-escape and virophage-first models and consider alternative scenarios regarding the origin of different lineages in order to gain a better understanding of their history,” says Postdoctoral Research Assistant José Gabriel Nio Barreat at the University of Oxford, UK.
The study was co-authored by Barreat and Aris Katzourakis, Professor of Evolution and Genomics at the Department of Biology at the University of Oxford.
Barreat and Katzourakis compared the nuclear escape scenario’s plausibility to that of other possible evolutionary scenarios by employing two hypothesis-testing strategies—maximum-likelihood and Bayesian frameworks. They zeroed in on four key proteins shared by infections in this genealogy that are associated with the arrangement of viral capsids: DNA-packaging ATPase, major and minor capsid proteins, and protease.
They applied two more techniques that utilized hereditary information to assess established phylogenies and induce the developmental direction of the various ancestries. After that, they looked into whether NCLDVs and adenoviruses shared a common ancestor, as predicted by the nuclear-escape scenario.
According to the nuclear-escape hypothesis, their analyses provided strong evidence against a sister relationship between NCLDVs and adenoviruses. To the exclusion of NCLDVs, the findings instead suggest that adenoviruses and mavericks shared a common ancestor. The researchers discovered that the most recent common ancestor of Mavericks and adenoviruses was not a virophage, which contradicts a virophage-first scenario. Nonetheless, their work doesn’t totally preclude the virophage-first speculation, making it the one best upheld by current phylogenetic analyses.
Moreover, their work offers help for the situating of the Bamfordvirae hereditary root among virophages and the other viral ancestries. The team discovered a novel model for these viruses’ evolutionary origins thanks to this positioning.
Aris Katzourakis, a co-author on the study, states, “The model proposes that the Bamfordvirae ancestor did not originate from an invasion of the eukaryotic cell nucleus and that it was a non-virophage DNA virus with a small genome.” Since virophages evolved into specialized NCLDV parasites, their way of life would have evolved later.
Katzourakis adds that the Bamfordvirae kingdom’s most recent common ancestor existed more than a billion years ago, including the earliest stages of eukaryotic life, based on the relative timing of events. However, there is currently no absolute timeframe for the precise dating of these events.
The deep divergences and extreme diversity in this lineage may have obscured the phylogenetic signal in the protein data analyzed, which is another limitation of the study. However, the focus on the viral capsid’s origin and development provides a straightforward explanation for the available data, and the authors were able to effectively differentiate between alternative scenarios.
According to Barreat, “This work contributes to our knowledge on how viruses evolve different evolutionary strategies, such as to become viral giants like NCLDVs or parasites of other viruses like virophages.” It is becoming increasingly apparent that viruses may have played a role in major evolutionary transitions throughout the history of life, in addition to playing important roles in Earth’s ecosystems. As a result, comprehending viruses’ extensive evolutionary history provides additional context for these ancient interactions and the participants.”
Katzourakis draws a conclusion by saying, “Understanding the interactions between viruses and their hosts provides a window into the deep evolutionary past that is illuminating the origins of both of these biological entities.”
More information: Jose Gabriel Nino Barreat et al, A billion years arms-race between viruses, virophages and eukaryotes, eLife (2023). DOI: 10.7554/eLife.86617.1