As a team with Southwest College, the State Key Lab of Silkworm Genome Science, and different accomplices, BGI Genomics has built a high-goal pangenome dataset addressing practically the whole genomic content of a silkworm.
This exploration paper, giving hereditary experiences into fake choice (taming and rearing) and biological variation, was distributed on September 24 in Nature Correspondences.
Already, because of the shortage of wild silkworms and the specialized limits of previous examinations, numerous quality-related locales were absent. This is the very first test to digitize silkworm genetic stock and make a “computerized silkworm”, incredibly working with practical genomic research, advancing exact rearing, and hence empowering extra silk use cases.
The group profoundly re-suscitated 1,078 silkworms (B. mori, including 205 nearby strains, 194 superior assortments, and 632 hereditary stocks and 47 wild silkworms, B. mandarina) and gathered long-read genomes on 545 of these examples, creating 55.57 T of genomic information.
“We intend to speed the process of silkworm molecular design breeding with thorough sampling and dataset combination with a variety of tests to uncover genes for future prospective study.”
Shuaishuai Tai, co-author and BGI Genomics senior researcher
This pangenome dataset contains the most complete data on the genomes of homegrown and wild silkworms and is the biggest long-perused pangenome on the planet for plants and creatures to date. Simultaneous top-to-bottom investigations on different hereditary varieties, population structure, phony choice, natural variations, and monetary qualities of silkworms have yielded fruitful results.
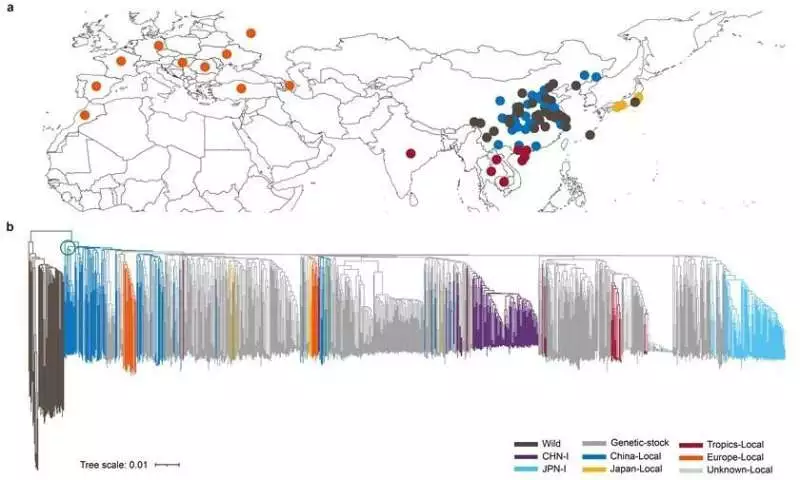
A Portrayal of the Primary Variety in 545 Silkworm Genomes BGI Genomics is to blame.
The beginnings of the homegrown silkworm
The homegrown silkworm, B. mori, was tamed from the wild mulberry silkworm, B. mandarina. It has a set of experiences that is north of 5,000 years old, yet its taming beginning area has for some time been an open inquiry because of an absence of solid natural proof.
The material in this study addresses the most extravagant hereditary variety from all significant sericulture areas of the world. The review figured out that endemic species from China’s lower and center Yellow Stream locales are conveyed at the foundation of the homegrown silkworm branch on the developmental tree, hence proposing that the homegrown silkworm began around here. The accessible archeological proof, including a half-case exhumed in 1926 at Xiyin Town, Xia Region, Shanxi Territory, and a stone-cut silkworm pupa unearthed in 2019 at Shicun in a similar province, offers significant help to this end.
Geographic conveyance and phylogenetic tree of the silkworm BGI Genomics is to blame.
Breaking the bottleneck in silkworm rearing
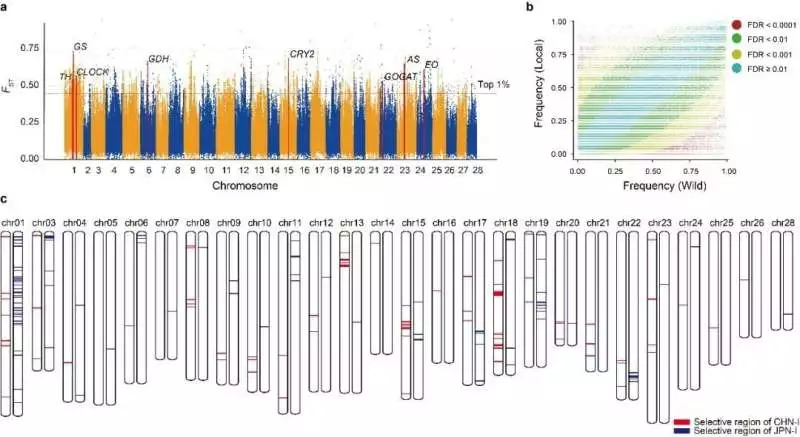
The customary rearing of silkworms has a long and novel history, yet since the 1990s it has stayed caught in a bottleneck. Precise examination of the hereditary premise of taming and improvement choice is fundamental to settling the annoying issues in silkworm rearing. The group has recognized 468 taming-related qualities and 198 improvement-related qualities, of which 264 and 185 have recently been distinguished. These characteristics will be important applicant focuses for silkworm atomic improvement.
Simultaneously, it was figured out that the Chinese and Japanese utility species share under 3% of the improvement loci. This not just reveals the somewhat free rearing accounts of Chinese and Japanese silkworms, but in addition, makes sense of why this common hereditary premise gives such mixed benefits to the two species. This result provides new knowledge for silkworm rearing in the future.
The monetary qualities of silkworm rearing
The yield and nature of silk have for some time been designated as the super monetary rules for the choice of silkworm. In any case, up to this date, there has been little significant awareness of how qualities and loci control these quantitative attributes. The pangenome appears to be the “closest scaffold” between aggregates, especially those with particularly complex properties.
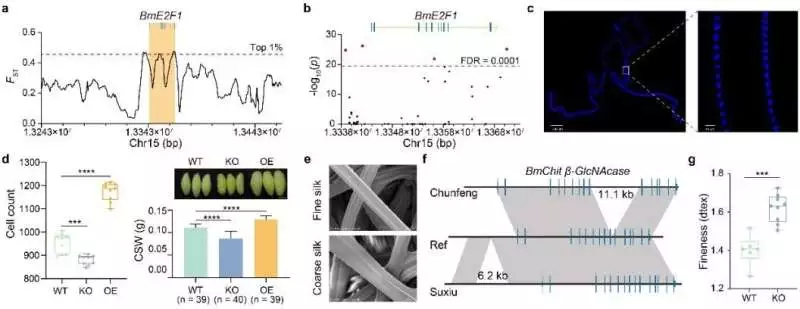
Silkworm taming and rearing BGI Genomics is to blame.
A valid example is the guideline of silk creation by the phone cycle-related record factor BmE2F1, which was uncovered through choice flagging and primary variety. CRISPR-cas9 interceded knockout of BmE2F1 reduces the quantity of silk organ cells by 7.68% and silk yield by 22%. Alternately, the transgenic overexpression of BmE2F1 increases the quantity of silk organ cells by 23% and silk yield by 16%.
Fine silk has novel applications and higher monetary worth, yet the hereditary premise of fiber fineness remains obscure. The identification of BmChit -GlcNAcase, a quality controlling silk fineness that can essentially be recognized in fine assortments, was prompted by examination of uncommon variations in the genomes of thin assortments, and CRISPR-cas9 intervened to knockout, resulting in coarser silk fineness created by homegrown silkworms.This proposes that this quality assumes a key part in deciding silk fineness.
The versatile qualities of silkworm rearing
Diapause is a typical natural versatile quality in bugs that guarantees that bugs can get by in spite of ominous ecological circumstances. Although the diapause chemical was first recognized in the silkworm in 1957, little data is accessible on the early stage diapause quality. In this review, in view of the examination of the pnd strain and genomic primary variety in the silkworm, and useful approval by quality altering, the BmTret1-like quality uncovered itself to be a significant determinant of post-undeveloped slowing down. This is the first occasion when a post-early stage determinant quality has been recognized in a bug.
This study uncovers the total dish genome of the silkworm to expose fake choice and natural variation experiences. Shuaishuai Tai, co-creator and BGI Genomics senior scientist, remarked, “With thorough testing and dataset joined with various trials to recognize qualities for future likely review, we desire to speed up the course of silkworm atomic plan rearing.”
More information: Xiaoling Tong et al, High-resolution silkworm pan-genome provides genetic insights into artificial selection and ecological adaptation, Nature Communications (2022). DOI: 10.1038/s41467-022-33366-x
Journal information: Nature Communications





