Princeton researchers discover the first route for selenium addition to normal items.
Princeton University researchers discovered a biosynthetic cycle that incorporates selenium into microbial small particles, indicating when such iotas were first discovered in natural products and opening up new avenues in selenobiology.
The discoveries likewise suggest that selenium, a fundamental minor component tracked down in all realms of life, may have a more significant natural job in microbes than researchers recently suspected.
The discoveries, which were distributed in the journal Nature, were written by Pursue Kayrouz, Jonathan Huang, Nicole Hauser, and Mohammad Seyedsayamdost.
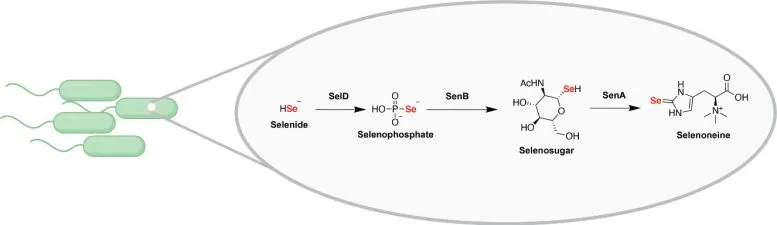
This outline shows the biosynthetic pathway integrating selenium into microbial little atoms, which highlights an undiscovered “compound space” in microorganisms that can now be dug for novel normal items. Credit: Nature
“This was a bit of a closed field.””No one had tracked down another pathway in selenium digestion in 20 years,” said Kayrouz. “The biosynthesis of selenoproteins and selenonucleic acids was clarified during the 1980s and 1990s. Also, from that point forward, individuals sort of accepted that these were the main things organisms did with selenium. We just contemplated whether they could integrate selenium into other little atoms. “They all come to an end.”
“In the 1980s and 1990s, the biosynthesis of selenoproteins and selenonucleic acids was clarified. Since then, it has been widely believed that these are the only uses of selenium by microorganisms. We merely questioned if they may add selenium to other tiny compounds. It appears that they do.”
Chase Kayrouz
Seyedsayamdost states, “Our work shows that nature has for sure developed advanced pathways to integrate this component into little atoms, sugars, and optional metabolites.” Selenium has amazing properties that are different from those of some other components found in biomolecules. For instance, the fusion of selenium into selenoneine gives it much better cell reinforcement than the sulfur variant of the particle. Yet, while sulfur is universal in biomolecules, selenium is a lot more uncommon and was believed to be restricted to biopolymers.
He proceeds, saying, “Nature has advanced explicit systems for integrating either sulfur or selenium into normal items, in this way exploiting the novel properties of the two components through pathways that are well defined for each.”

Looking for Selenium
The analysts started their examination with the suspicion that selenium iotas ought to happen in normal items because of their widespread use elsewhere. They considered what such a mark could resemble in microbial genomes.
“How would you know where another medication, normal item, or selenium metabolite is, and how would you track it down?” Kayrouz asked. “We normally search for biosynthetic quality bunches—gatherings of qualities on the chromosome that code for the biosynthesis of such atoms.” Thus, in the event that we have a pathway to make a selenium-containing compound, it must be encoded by qualities.
They executed a genome mining system looking for qualities that are viewed as close to selD, which encodes the most vital phase in fully realized selenium processes inside the cell.
They found one quality that was co-limited with selD—called senB—that caught their attention, particularly because it had not previously been ensnared in selenium digestion.
SenA, a third co-limited quality, was discovered through further investigation. Kayrouz guessed that these three qualities might be engaged with another selenium biosynthetic pathway.
“To start with, we characterized what a biosynthetic-quality bunch that integrates selenium would seem to be,” said Seyedsayamdost. “We then utilized bioinformatics to search for such qualities and recognized what we presently call the “sen group” in assorted microbial genomes.”
They had the option of communicating every one of these new qualities to Escherichia coli, hence gathering the whole pathway in a test tube. This uncovered the creation of two selenium-containing little particles: a selenosugar and an atom called selenoneine. It likewise uncovered two proteins that structure carbon-selenium bonds, the main catalysts to follow up on natural little atoms.
“The organisms are placing selenium into these mixtures for an explanation, so there should be some intriguing bioactivity related to them,” said Kayrouz. “We don’t have any idea what that is yet, yet it is very energizing. “As natural scientists, disclosures like this are what we awaken for each day.”
Reference: “Biosynthesis of selenium-containing little atoms in assorted microorganisms” by Pursue M. Kayrouz, Jonathan Huang, Nicole Hauser, and Mohammad R. Seyedsayamdost, 7 September 2022, Nature.
DOI: 10.1038/s41586-022-05174-2





