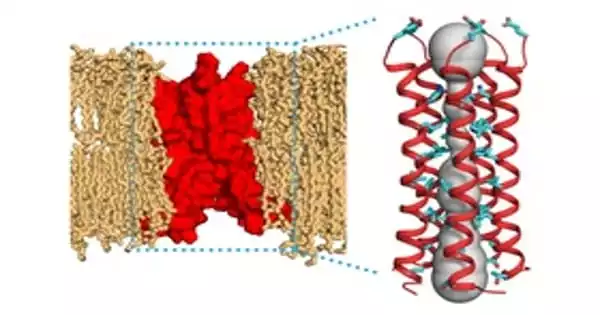
Chemists have found the structure of a protein capable of pumping harmful chemicals out of bacterial cells. Knowing this structure could lead to the development of medications that block transport proteins and help resensitize drug-resistant bacteria to existing antibiotics.
The structure of a protein that can pump hazardous chemicals out of bacterial cells has been identified by MIT researchers. Proteins like this one, discovered in E. coli, are thought to help bacteria grow resistant to a variety of antibiotics.
The researchers were able to detect how the structure of this protein changes as a drug-like molecule travels through it by using nuclear magnetic resonance (NMR) spectroscopy. According to Mei Hong, an MIT professor of chemistry, knowing this particular structure could lead to the development of medications that can inhibit these transport proteins and help resensitize drug-resistant bacteria to existing antibiotics.
“With knowledge of the structure of this protein’s drug-binding pocket, one could try to develop competitors to these substrates, so that you could block the binding site and prevent the protein from removing antibiotics from the cell,” says Hong, the paper’s senior author.
Alexander Shcherbakov, an MIT graduate student, is the study’s principal author, and it was published today in Nature Communications. Aurelio Dregni, an MIT graduate student, and Katherine Henzler-Wildman, a professor of biochemistry at the University of Wisconsin at Madison, are also members of the research team.
The SMR transporters have substantial sequence conservation throughout critical areas of the protein. EmrE is by far the most researched member of the family, both in vitro and in vivo, making it an appropriate model system for studying the structure that enables SMR activity
Katherin Henzler-Wildman
Transporters of drug resistance
One of the various techniques bacteria can try to avoid medicines is to pump chemicals out through their cell membranes. Henzler-group Wildman’s at the University of Wisconsin has been studying EmrE, a membrane-bound protein that may transport a variety of hazardous chemicals, including herbicides and antimicrobial agents, for several years.
EmrE is a member of a protein family known as small multidrug resistance (SMR) transporters. Although EmrE is not directly implicated in antibiotic resistance, other members of the family have been detected in drug-resistant strains of Mycobacterium TB and Acinetobacter baumanii.
“The SMR transporters have substantial sequence conservation throughout critical areas of the protein. EmrE is by far the most researched member of the family, both in vitro and in vivo, making it an appropriate model system for studying the structure that enables SMR activity” Henzler-Wildman explains.
A few years ago, Hong’s lab developed a technique that allows researchers to use NMR to measure the distances between fluorine probes and hydrogen atoms in proteins. This makes it possible to determine the structure of a protein as it binds to a molecule that contains fluorine.
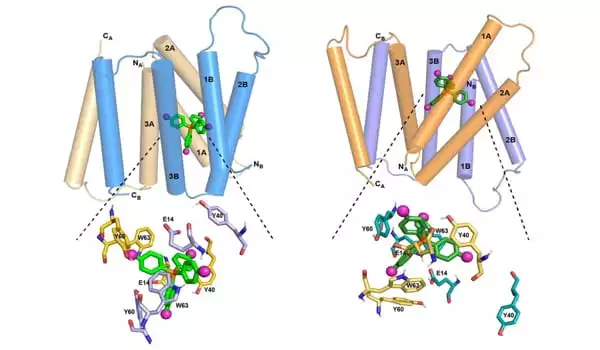
After seeing Hong speak about the new technique at a convention, Henzler-Wildman recommended they collaborate to explore EmrE. Her lab has spent several years researching how EmrE carries a drug-like molecule, known as a ligand, across the phospholipid membrane. This ligand, F4-TPP+, is a tetrahedral molecule with four fluorine atoms linked to each corner.
The researchers set out to discover the atomic-resolution structure of EmrE using this ligand and Hong’s innovative NMR approach. Each EmrE molecule already has four transmembrane helices that are fairly parallel. Two EmrE molecules form a dimer, and eight transmembrane helices form inner walls that interact with the ligand as it passes through the channel. Previous research revealed the overall structure of the helices, but not the protein side chains that extend into the channel interior and act as arms to grab the ligand and guide it through the channel.
EmrE carries harmful chemicals from the neutral pH inside a bacterial cell to the acidic pH outside. The pH shift across the membrane has an effect on the structure of EmrE. Hong and Henzler-Wildman discovered the structure of the protein as it interacts to F4-TPP+ in an acidic environment in a publication published in 2021. They studied the structure at a neutral pH in the current Nature Communications paper, allowing them to determine how the protein structure changes when the pH varies.
A complete structure
The researchers discovered in this study that at neutral pH, the four helices that make up the channel are relatively parallel to one another, producing an aperture through which the ligand may easily enter. As the pH decreases and moves closer the membrane’s edge, the helices begin to tilt, making the channel more open to the outside of the cell. This assists in pushing the ligand out of the channel. At the same moment, many rings in the protein side chains adjust their orientation, assisting in the ligand’s exit from the channel.
The acidic end of the channel is also more inviting to protons, which enter the channel and aid in its opening, allowing the ligand to depart more easily. “This study really brings the story to a close,” Hong says. “One structure is insufficient. You’ll need two to figure out how a transporter can open to both sides of the membrane, because it’s designed to push the ligand or antibiotic component from inside the bacteria out.”
Because the EmrE channel is thought to convey a wide range of harmful substances, Hong and her colleagues intend to investigate how additional molecules pass through it. The National Institutes of Health and the MIT School of Science Camplan Fund contributed to the study’s funding.