A new study by bioscientists at Rice University explains how a model plant organism’s basic cellular structures work together to fuel growth. The discoveries could reveal insight into comparing systems in human cells.
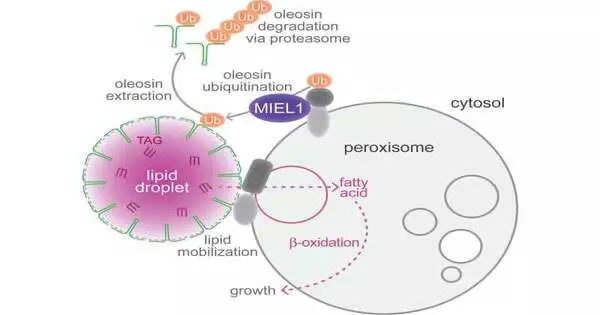
Before it is sufficiently grown to perform photosynthesis, an Arabidopsis thaliana seedling depends on the hold of fats put away inside its cells in protein-covered pockets known as lipid beads. The energy-rich items in these beads are prepared in intracellular holders called peroxisomes. According to a new study, this collaboration necessitates the presence of an enzyme on the peroxisome that aids in the breakdown of the protein coating on the lipid droplets. In the past, it was known that the enzyme MIEL1 was found in the cell nucleus, where it helped control gene expression.
The disclosure of another job and area for MIEL1 provoked questions about whether or not the discoveries would likewise apply to its human partner, PIRH2. The study, which was published in the Proceedings of the National Academy of Sciences, found that PIRH2 associates with peroxisomes when expressed in Arabidopsis cells after additional experiments. A deeper comprehension of the cellular function of PIRH2 could provide insights into cancer prevention and treatment due to its significant role in tumor formation.
“PIRH2 is one of the most well-studied p53 regulators, so it’s directly linked to cancer, because p53 is frequently mutated and implicated in a wide range of cancers in a variety of organs and cell types.”
Melissa Traver, a postdoctoral research associate in the Bartel lab who is the lead author on the study.
PIRH2 plays a role in the degradation of p53, a well-known protein that regulates the proliferation of cells with damaged genomes in human cells. The “guardian of the genome” gene that encodes p53 has been extensively studied for its ability to suppress tumor growth. Transformations that disturb its capability are tracked down in many tumors.
“PIRH2 is one of the most profoundly considered of p53 controllers, so it’s straightforwardly attached to disease, in light of the fact that p53 is many times transformed and embroiled in various tumors in a variety of organs and cell types,” said Melissa Traver, a postdoctoral exploration partner in the Bartel lab who is the lead creator on the review.
“Hence, it’s an intriguing quality to study,” added Traver, who is likewise a Rice alum of the natural chemistry and cell science doctoral program. “Diving more deeply into it could illuminate what we are familiar with: the way in which malignant growth works and how to stop it. I never felt that I would wind up perusing papers about disease in a plant lab. I began this task attempting to answer an extremely explicit inquiry regarding plants, and it has been very reassuring and rewarding to find something possibly more broadly pertinent across frameworks.”
The discoveries supported Traver’s conviction that crucial exploration can yield influences as significant as application-based science.
According to Traver, “I’ve spent a lot of time defending basic science and stressing its necessity and intrinsic value” as a graduate student.

Packets of Arabidopsis thaliana seeds are examined by Melissa Traver. Credit: Jeff Fitlow/Rice College
Along with Bonnie Bartel, Rice’s Ralph and Dorothy Looney Teacher of BioSciences, Traver inspected the physical processes that unfold during Arabidopsis germination.
“For a really long time, we’ve been contemplating peroxisomes in Arabidopsis—how they’re made, what they do, and for what reason they’re significant,” said Bartel. “Peroxisomes assume a particularly basic role during germination, when the plant goes through huge development but isn’t yet adequately experienced to do photosynthesis. This implies that it needs to utilize the lipids put away in the seed by the parent plant.
“The close association of these two organelles, one where fats are stored and one where they are processed, sparked our interest in lipid droplets. Lipid drops have a protein covering that holds them back from blending with each other. We were interested in the method by which oleosin, a protein, is eliminated from cells.
Traver engineered a protein that was tagged with a fluorescent marker to learn how oleosin breaks down.
“We are geneticists, so when we need to comprehend something, we like to break it,” Bartel said. ” Melissa chose to search for plants that couldn’t debase this oleosin in the manner that the wild kind does. She could see that the wildtype seedling was initially lit up due to the fluorescent tag on the oleosin. However, as the lipids and oleosin are consumed, the fluorescence diminishes.”
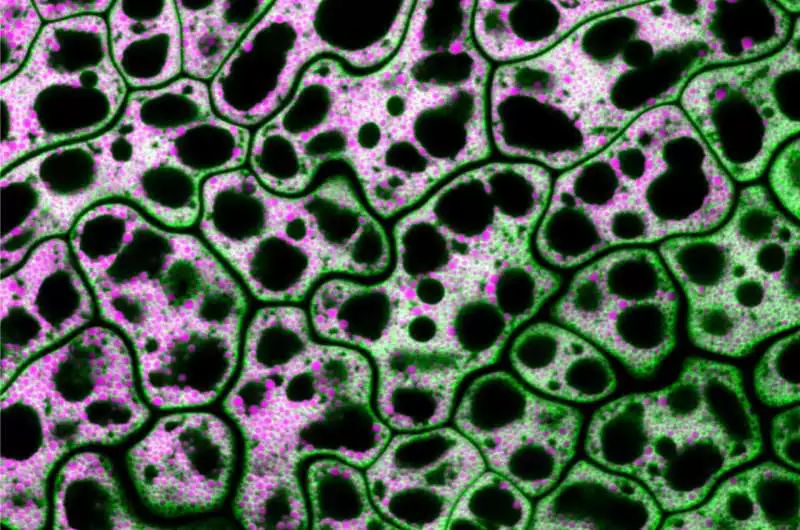
Envisioned are cells from a MIEL1-freak Arabidopsis seed with unbiased lipids displayed in fuchsia and a lipid drop coat protein displayed in green. The dynamic collaboration between lipid droplets and peroxisomes, organelles that store and catabolize fats, was investigated by Traver and Bartel using genetics and microscopy. Credit: Melissa Traver/Rice University.
Conversely, the freak seedlings that can’t separate oleosin keep showing fluorescence. Traver was successful in locating the gene that is responsible for the plants’ capacity to degrade oleosin by sequencing their genome and comparing it to that of the wild-type plant.
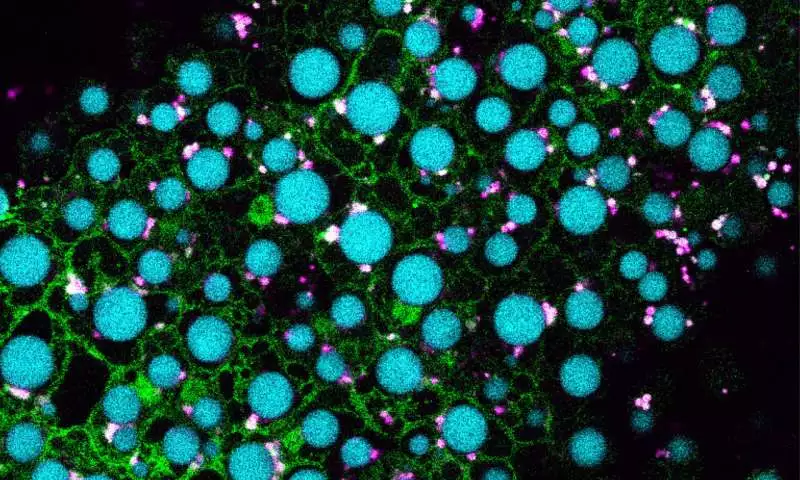
Pictured are cells from an Arabidopsis seedling overexpressing MIEL1 (green) with lipid droplets shown in cyan and peroxisomes shown in magenta. Traver and Bartel used genetics and microscopy to explore the dynamic collaboration between lipid droplets and peroxisomes, organelles that store and catabolize fats, respectively. Their evidence suggests that MIEL1 promotes degradation of coat proteins on lipid droplets adjacent to peroxisomes. Credit: Melissa Traver/Rice University.
“The quality that is done in the freak seedlings codes for MIEL1, an atomic compound that debases record factors—proteins that balance quality articulation,” Bartel said.
Traver conducted additional experiments to determine whether MIEL1 is associated with peroxisomes or lipid droplets.

Traver (left) and Bartel discovered that peroxisomes, the specialized intracellular structures where plant cells process lipids, are accompanied by an enzyme that breaks down the protein coating of lipid droplets, lipid-storing organelles. Credit: Jeff Fitlow/Rice University
“The startling thing Melissa found is that despite the fact that it follows up on lipid beads, MIEL1 is really restricted at the peroxisome,” Bartel said. “Lipid beads and peroxisomes are dissipated all through the cell, and you would rather not corrupt the oleosin except if a lipid drop is right close to a peroxisome. Our speculation is that having this chemical on the peroxisomes is a method for guaranteeing the right natural chemistry happens right where it’s required.”
The review’s discoveries propose that the compound-mediated cooperation among peroxisomes and lipid beads could seem to be comparative across all eukaryotic living things.
Traver stated, “The next thing to do would be to carry out experiments in human cells or other animal models to see if similar mechanisms are at play.”
More information: Melissa S. Traver et al, The ubiquitin-protein ligase MIEL1 localizes to peroxisomes to promote seedling oleosin degradation and lipid droplet mobilization, Proceedings of the National Academy of Sciences (2023). DOI: 10.1073/pnas.2304870120