Biomedical specialists at Duke College have fostered an artificial intelligence stage that independently thinks about particles and gains from their varieties to expect property contrasts basic to finding new drugs. The stage gives specialists a more exact and effective instrument to help plan therapeutics and different synthetic compounds with helpful properties.
The examination was distributed on October 27 in the Diary of Cheminformatics.
AI calculations are progressively used to study and foresee the organic, compound, and actual properties of little atoms utilized in drug advancement and other material planning errands. These instruments can assist scientists with figuring out the key “ADMET” properties of a particle—how it’s retained, dispersed, processed, discharged, and its poisonousness inside the body. By understanding these various properties, analysts can distinguish particles to foster new therapeutics that are more secure and more viable.
“By instructing the network to learn from a one-to-one comparison, you provide it with more data points than if it were learning from one molecule at a time. The platform learns about the structure and properties of each molecule independently, but it also learns about the differences between the two and how those differences influence the qualities of the molecule.”
Reker and Zachary Fralish, a Ph.D.
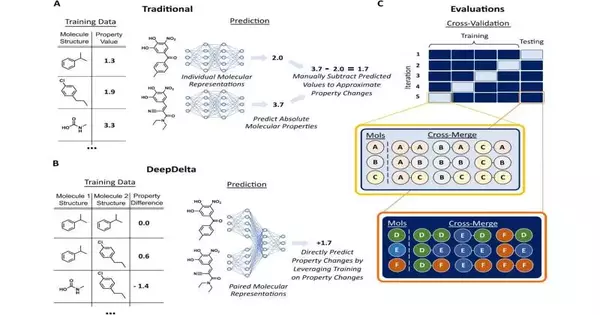
While existing AI stages empower scientists to screen a much larger number of particles than would be conceivable by genuinely making them all in a lab, they can foresee the properties of each particle in turn, restricting their general proficiency when entrusted to recognize the most ideal compound.
While there are a couple of other computational ways to deal with this additional step and straightforwardly look at particles, they are restricted in their degree. For instance, techniques like free energy bother are exceptionally precise yet so computationally complex that they can assess a modest bunch of particles all at once. Approaches like matched atomic matches, then again, are a lot quicker but can look fundamentally the same as particles, restricting their more extensive use.
To resolve this issue, Reker and Zachary Fralish, a Ph.D. understudy in the Reker lab, created DeepDelta, a profound learning approach that can productively look at two particles at the same time and foresee the property distinctions between them regardless of whether they are totally different.
“By having the organization gain from a balanced correlation, you’re giving it a bigger number of data points of interest than if it were gaining from each particle in turn,” said Reker. “The stage is finding out about the design and properties of every atom separately, but on the other hand, it’s finding out about the distinctions between the two and how those distinctions illuminate the particle’s properties.”
The group tried the DeepDelta stage against two cutting-edge models in the field: Irregular Backwoods, a broadly utilized exemplary AI model, and ChemProp, a profound brain network that DeepDelta is dependent on. Every framework analyzed two known atomic designs and anticipated 10 different ADMET properties, including how the particles are cleared from the kidneys, their particular half-lives, and how well they can be used by the liver.
DeepDelta demonstrated that it was essentially more viable and exact at foreseeing and measuring the distinctions in sub-atomic properties between particles than the current stages.
“Preparing on sub-atomic contrasts permits this strategy to be more precise while choosing if another synthetic is preferred or more awful over an ongoing one,” said Fralish. “Like doing schoolwork, it is more similar to your test. We additionally significantly extended the size of our datasets by matching, basically giving our models more schoolwork, which truly assists information-hungry brain networks with finding out more.”
The group is currently anticipating integrating this model into their work as they plan likely new treatments and advance existing medication competitors.
“With this device, we could take a gander at a medication that nearly endured FDA endorsement; however, perhaps it definitely disapproved of liver harmfulness, so it didn’t exactly make it,” said Fralish. “DeepDelta could assist with distinguishing particles that have those equivalent great properties yet no liver poisonousness. This device opens up a ton of chances by assisting us with concluding what substance has the most obvious opportunity with regards to doing what we need in reality, setting aside time and cash.”
More information: Zachary Fralish et al, DeepDelta: predicting ADMET improvements of molecular derivatives with deep learning, Journal of Cheminformatics (2023). DOI: 10.1186/s13321-023-00769-x