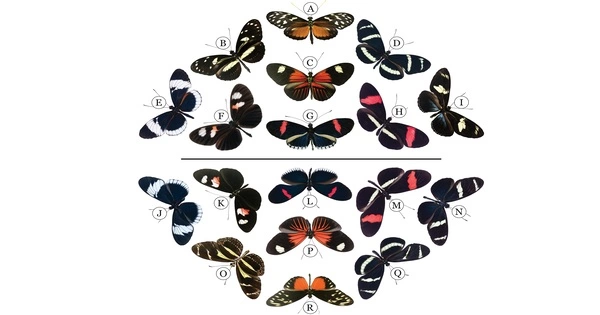
The Nymphalidae are the largest family of butterflies, with over 6,000 species found all over the world. They are medium-sized to large butterflies that belong to the superfamily Papilionoidea. Most species have shortened forelegs, and many rest with their colorful wings flat.
The use of only four legs is the most well-known feature of these butterflies; the reason their forelegs have become vestigial is unknown. Because some species have a brush-like set of soft hair called setae, researchers believe the forelegs are used to improve signaling and communication between the species, while standing in the other four. This ability is beneficial to reproduction and the overall health of the species, and it is currently the leading theory.
The cis-regulatory evolution of the gene WntA in nymphalid butterflies has been described by a team of researchers from Cornell University and The George Washington University. The researchers used a variety of techniques in their study, which was published in the journal Science, to better understand how gene regulatory processes in a type of butterfly can allow for both deep homology and rapid adaptation to environmental changes.
Prior research has shown that patterns in the appearance of creatures such as butterflies and the structures behind them arise due to the influence of transcription factors and cis-regulatory elements (CREs.) But how such factors have evolved is still not very well understood. In this new effort, the researchers sought to gain insight into the regulatory systems that underpin rapidly evolving traits.
Marianne Espeland and Lars Podsiadlowski with Leibniz Institute for the Analysis of Biodiversity Change at Museum Koenig, have published a Perspective piece in the same journal issue outlining the means by which gene regulatory agents play a major role in the pattern formation of butterfly wings and the work done by the team in this new effort.
Prior research has shown that patterns in the appearance of creatures such as butterflies and the structures behind them arise due to the influence of transcription factors and cis-regulatory elements (CREs.) But how such factors have evolved is still not very well understood. In this new effort, the researchers sought to gain insight into the regulatory systems that underpin rapidly evolving traits.

To learn more about such systems, the researchers conducted comparative sequence analysis, ATAC-seq on five species of butterflies, focusing specifically on 46 CREs and the WntA gene. They also used CRISPR knockouts to learn more about the role that the WntA gene plays in wing color. They found strong evidence that suggests regulatory genetic elements from the deep evolutionary past in nymphalid butterflies exert a strong influence on the ways that patterns have developed on their wings.
The researchers found that the WntA gene is a master gene, playing a critical role in the development of patterns that appear on butterfly wings. They found that its expression has an impact not just on color and patterns, but on the expression of several other genes located near it in the butterfly DNA.
And by generating deletions using CRISPR-Cas9, they were able to study the impact of multiple transcription factors and CREs on wing pattern and color. They found multiple changes in one species that were not evident in others, suggesting newly evolved CREs that had very quickly become part of the regulatory process.