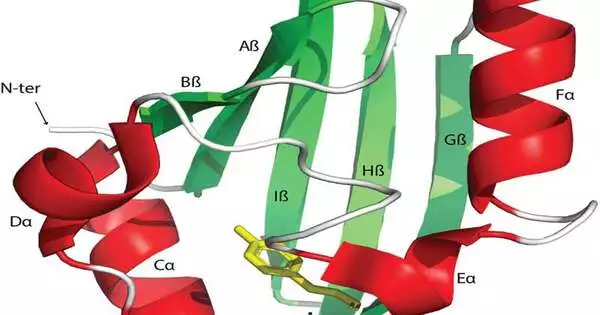
Signal transduction and discernment manage natural exercises to adjust to evolving conditions. The Sprightly Arnt-Sim spaces are normally accessible sensors tracked down across assorted receptors in microorganisms, eukaryotes, and archaea. Be that as it may, the degree of their utilitarian variety and their dispersion across the tree of life still need to be described.
In another report in Science Advances, Jiawei Xing and a group of researchers in science and translational information at the Ohio State College, U.S., have utilized succession protection and primary data to propose explicit and possible capabilities for almost 3,000,000 energetic Arnt-Sim spaces (contracted as PAS areas). The work recommended the beginning of these areas in microscopic organisms and their even exchange with archaea and eukaryotes. The cozy relationship of the spaces among people and microorganisms gives an exceptional chance to drive a drug plan.
The impact of sensor spaces on natural pathways
Signal transduction pathways are focal roads in all living cells that distinguish supplements, chemicals, oxygen, and redox potential to control different cell capabilities. These signs can be perceived by receptor proteins by utilizing committed sensor areas. The PAS spaces are found in record factors, protein kinases, catalysts controlling courier turnover, and chemoreceptors. In spite of the fact that sensor spaces are normally extracytoplasmic, these areas are predominantly cytoplasmic. Additionally, the PAS spaces present in human record factors are key medication focuses for disease treatment.
In this work, Xing et al. give a broad relative genomic examination of PAS areas across microscopic organisms, archaea, and eukaryotes. The group showed the starting points of PAS spaces in microorganisms and how their tactile capabilities advanced in various ways. The eukaryotes obtained PAS spaces from microorganisms through an assortment of free quality exchange occasions.
The comparability of human PAS spaces to those in microorganisms suggests the usefulness of these proteins as valuable models to decide signal particularity relative to human partners. These results prepare to investigate utilitarian examinations and medication improvement in PAS spaces while filling in as a system to concentrate on protein families with different and flexible capabilities.
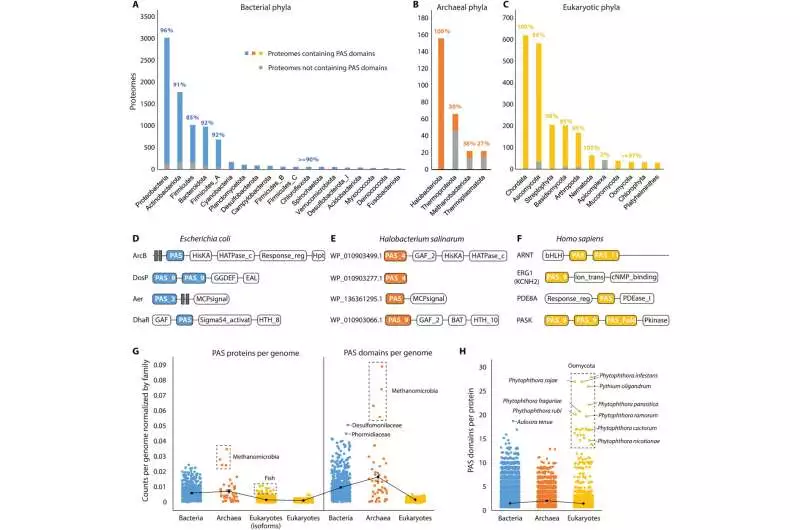
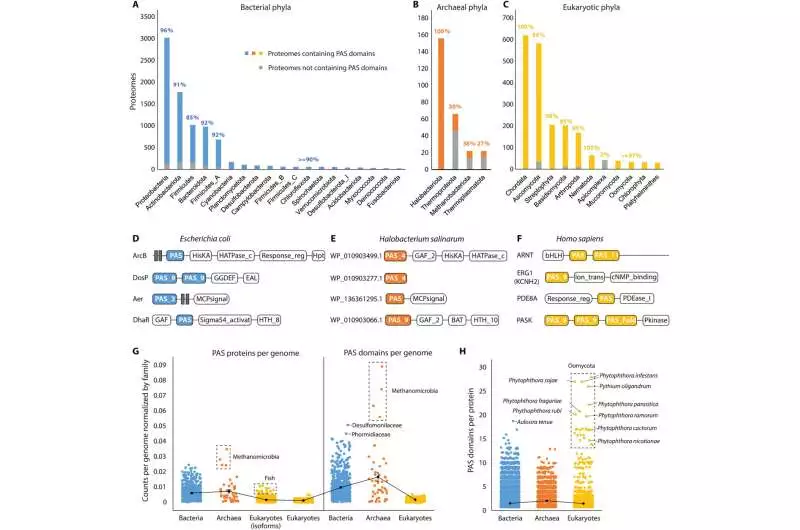
Worldwide appropriation of PAS areas (A to C) PAS space dispersions in reference proteomes from bacterial (A), archaeal (B), and eukaryotic (C) phyla Shaded bars show quantities of proteomes with PAS areas in every phylum. Dim bars show quantities of proteomes without PAS areas in every phylum. Rates show the extent of PAS-containing proteomes in every phylum. Phyla with less than 20 reference proteomes are not shown. (D to F) Agent PAS-containing proteins in three model living beings (G) Normal quantities of PAS proteins per genome (left) and PAS spaces per genome (right) across three realms (standardized by complete quality numbers, see Materials and Strategies). Each spot addresses the typical count of a family. (H) Quantities of PAS spaces per protein across three realms Each spot addresses the number of PAS spaces in a protein. Credit: Science Advances, doi: 10.1126/sciadv.adi4517
PAS space circulation across the tree of life
Since initially finding the PAS superfamily, the conveyances of the spaces have been deliberately examined across the tree of life. Xing and partners looked for these spaces in the whole arrangement of reference proteomes to lay out the presence of the area in 66% of archaeal, 93% of bacterial, and 93% of eukaryotic proteomes. The areas were generally present in bacterial phyla, unevenly conveyed in archaea, and broadly circulated in eukaryotes.
The researchers gathered all PAS-containing proteins from the InterPro data set and recognized their area structure. While histidine kinases are the most well-known PAS proteins in microscopic organisms and archaea, record factors are more accessible in eukaryotes. The archaeal and bacterial genomes encode a greater amount of proteins and spaces than eukaryotic genomes.
The current order of PAS spaces doesn’t mirror their natural capabilities.
The present Pfam data set of protein families organizes the PAS area into 17 families. To investigate, assuming that the current grouping of PAS spaces mirrors their natural capability, Xing et al. directed a complete writing search. The work showed that the current characterization of PAS areas doesn’t reflect tactile capabilities.
The group likewise directed an impact search utilizing successions from families. They followed up these analyses by distinguishing PAS families utilizing the succession and primary data from the NCBI reference grouping information base, as well as the whole PFAM data set.
Utilitarian bunches of PAS areas (A) Bunching results Potential capabilities represent arrangements with protection, however obscure. Explicit capabilities show successions with anticipated cofactors. Quantities of arrangements in every classification are shown. (B) Markov groups of PAS areas Bunches are numbered by the number of successions (groups with more modest numbers contain more arrangements). Shaded edges interfacing groups reflect shooting hits between bunches (yellow to red from not many to many). Just bunches 1 to 163 with moderate buildups inside the hole are shown. Anomaly bunches are not displayed for effortlessness. Bunches with explicit monitored cofactors are featured: blue, heme restricting; orange, Prevailing Fashion restricting; red, FMN restricting; yellow, PYP homologs. Delegate proteins are marked close to these bunches. (C) Cofactor-restricting designs A delegate PAS space is displayed for each bunch: group 3, FixL (PDB: 1DRM); bunch 119, Aer2 (PDB: 4HI4); bunch 144, PYP (PDB: 2PHY); group 4, NifL (PDB: 2GJ3); bunch 14, Aer (PDB: 8DIK); bunch 11, Phot1 (PDB: 2Z6C). Yellow, cofactor; green, monitored deposits for cofactor restriction. (D) Conveyance of PAS area bunches The size of the diagram addresses the number of PAS areas. Colors show PAS areas for various proteins. HK/RR, two-part frameworks; GGDEF/EAL, cGMP controllers. Credit: Science Advances, doi: 10.1126/sciadv.adi4517
Key discoveries
Xing and partners showed that these spaces have short and different arrangements that muddled their phylogenetic examination. Be that as it may, the spaces had a similar primary crease. The group examined 1% of successions from each bunch and developed phylogenetic trees by means of a construction-directed approach for short and various arrangements.
They noticed the succession, underlying, and phylogenetic data to propose that PAS spaces in such groups autonomously advanced the abilities for heme-restricting. The concentrate likewise uncovered the eukaryotic PAS spaces to have bacterial starting points and the ability to offer promising medication targets.
Viewpoint
Along these lines, the exploration group directed a vast study of the PAS (Saucy Arnt-Sim) spaces 24 years after the latest investigation. In this work, the group dissected almost 3 million PAS spaces from the huge genomes of microscopic organisms, archaea, and eukaryotes. The specialists introduced their discoveries about advancement, the genomic scene, and the utilitarian expansion of organic sensors. The results demonstrated a few PAS spaces to be available in an assortment of living things while likewise recognizing them in significant sorts of sign transduction proteins.
Since the current characterization frameworks are obsolete to a degree, Xing and his group utilized a few techniques to deliver a superior order framework in this work. The review uncovered the PAS spaces as general sub-atomic sensors for configuration-designated tests for approval and further review.
More information: Jiawei Xing et al, Origin and functional diversification of PAS domain, a ubiquitous intracellular sensor, Science Advances (2023). DOI: 10.1126/sciadv.adi4517
Gregg L. Semenza, Hypoxia-Inducible Factors in Physiology and Medicine, Cell (2012). DOI: 10.1016/j.cell.2012.01.021