The remaining noncoding proteins, which most likely aid in gene regulation, make up less than 2% of the human genome. Transformations in the noncoding genome frequently trigger attribute changes that cause illness or handicap by adjusting quality articulation. Notwithstanding, it tends to be difficult for researchers to find which of the various variations related to a sickness or other complex quality are the causal ones and to figure out the system of their belongings.
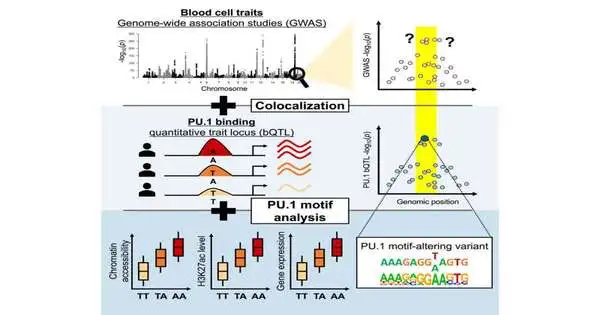
Specialists at the Brigham and Women’s Hospital fostered another computational methodology that focuses on little locales of the noncoding genome that expansive affiliation studies (GWAS) distinguished as being related to changes to platelet qualities, including decreased lymphocyte counts and hemoglobin fixations.
They then filtered these districts for explicit changes that caused a record factor protein, called PU.1, to tie to specific regions pretty much unequivocally than ordinary and inspected the impact that such transformations had on PU.1’s limiting site. 51 of the 69 mutations that altered PU.1’s binding site and were associated with quantitative differences in blood cell trait changes were identified using their method, suggesting that they probably caused a physiological difference.
“Our approach could be used to elucidate a variety of genetic disorders and to hone in on the causal variants in the noncoding genome underlying different biomedical traits,”
Senior author Martha Bulyk, Ph.D., a Principal Investigator in the Brigham’s Division of Genetics.
“Our technique could be applied to all the more likely grasp a scope of hereditary circumstances and to assist with pinpointing the causal variations in the noncoding genome’s basic biomedical qualities,” said senior creator Martha Bulyk, Ph.D., a vital examiner in Brigham’s Division of Hereditary Qualities.
“In this case, we found noncoding variants that appear to be associated with quantitative differences in the changes in blood cell trait.” The transcriptional regulatory mechanisms hidden in the GWAS data of other complex traits could be discovered using this strategy.”
The study has been published in the Cell Genomics journal.
More information: Raehoon Jeong et al, Blood cell traits’ GWAS loci colocalization with variation in PU.1 genomic occupancy prioritizes causal noncoding regulatory variants, Cell Genomics (2023). DOI: 10.1016/j.xgen.2023.100327