Our phones play out a wonder of design with regards to pressing data into little spaces. Each time a cell partitions, it groups up an astounding 4 meters of DNA into 46 little bundles, every one of which is just a few millionths of a meter long. Specialists from EMBL Heidelberg and the Julius-Maximilians-Universität Würzburg have now found how a group of DNA engine proteins prevails with regards to bundling inexactly organized strands of DNA into reduced individual chromosomes during cell division.
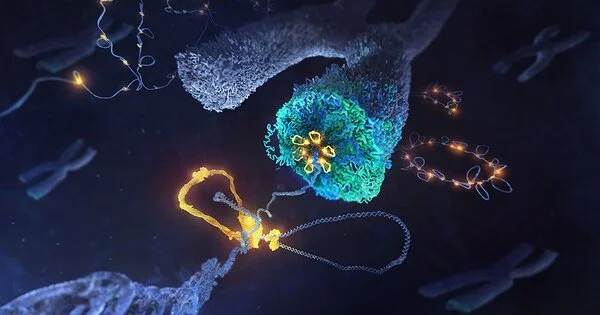
The specialists considered condensin, a protein complex basic to the course of chromosome development. Although this complex was found over thirty years ago, its method of activity remained generally neglected. In 2018, specialists from the Häring bunch at EMBL Heidelberg and their partners showed that condensin particles make circles of DNA, which might make sense of how chromosomes are shaped. The internal functions of the protein complex that allow it to accomplish this feat remained unknown.
“We’ve been working on this issue for quite some time. But it is only now that we have found an answer to this long-standing question by integrating multiple experimental methodologies.”
Christian Häring, former Group Leader at EMBL Heidelberg
“We have been dealing with this issue for quite a while. Yet, just now, by joining different exploratory methodologies, we have tracked down a response to this well-established question, “said Christian Häring, previous Group Leader at EMBL Heidelberg and presently Professor at the Julius-Maximilians-Universität Würzburg.
Through carefully planned tests, some of which included noticing and controlling single condensin particles while they were currently shaping DNA circles, the scientists found how various pieces of the complex all in all go about as a sub-atomic machine: one section holds the DNA consistent, similar to an anchor, while the other section goes about as an engine which pushes the DNA ahead, in this way making a wide circle.
Like other engine proteins, condensin takes “ventures” along the DNA, consuming cell energy as ATP at the same time. In any case, these means are in excess of quite a bit longer than the means taken by other DNA engine proteins, despite the fact that how much energy is utilized is generally something very similar. “It resembles a recipe for one dashing vehicle with the energy proficiency of an e-bicycle,” said Indra Shaltiel, the primary creator.
Headways in cryo-electron microscopy methods permitted us to picture this complicated system in extraordinary detail,” said Sebastian Eustermann, Group Leader at EMBL Heidelberg and a senior creator of the review distributed in Science. “We were able to capture condensin in real life and determine a sub-atomic movement of how ATP energizes its engine action — an important step toward understanding DNA helical development.” Comparative circles and related atomic machines have been embroiled in assorted genomic processes, including the control of how qualities are turned here and in the middle between cell divisions. Thusly, our discoveries might have significantly more extensive ramifications.
Condensins share a place with perhaps the most developmentally ancient groups of chromosomal proteins. The revelation of this new instrument hence opens up an entirely different field of study. “Individuals from the class of engine proteins condensin has a place with are probably fundamental for all life on the planet,” Häring said. “It is clear we are just barely beginning to comprehend their jobs and what they may be meant for in human circumstances.”