A huge amount of time is expected to decide the medication weakness profile of a bacterial disease. Presently, scientists from the Nara Foundation of Science and Innovation and their teaming up accomplices have developed an innovation that will decisively accelerate this generally sluggish cycle and perhaps assist in saving lives.
The CDC states that anti-toxin safe diseases are liable for killing north of 1,000,000 individuals overall consistently. The key to overseeing safe diseases is rapidly recognizing the proper treatment to which the infective microbes are helpless. “As a rule, weakness results are required a lot quicker than regular tests can convey them,” says Yaxiaer Yalikun, senior creator. “To address this, we cultivated an innovation that can address this issue.”
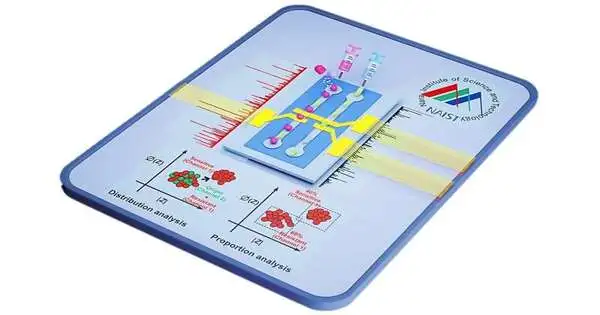
The gathering’s work depends on impedance cytometry, which estimates the dielectric properties of individual cells with a high throughput — in excess of 1,000 cells per moment. Since the electrical readout of a bacterium relates to its actual reaction to an anti-toxin, one has a clear method for deciding if the anti-toxin neutralizes the microbes.
“Although there was a less than 10% misidentification error in our work, there was a strong differentiation between susceptible and resistant cells within 2 hours after antibiotic treatment.”
Yoichiroh Hosokawa, another senior author in the group.
Regular impedance cytometry includes examining the test (anti-toxin treated) and reference (untreated) particles in a single example followed by aligning the impedance of the two particles. The two stages require specialized experts to do broad post-handling, which was a significant limit to survive.
In a review published in ACS Sensors, the gathering fosters a clever impedance cytometry strategy that all the while dissects the test and reference particles into isolated channels, making effectively analyzable separate datasets. This cytometry had nanoscale awareness, considering the location of even momentary actual changes in bacterial cells.
In a simultaneous report distributed in Sensors and Actuators B: Compound, the gathering planned an AI device to examine the impedance cytometry information. Since the new cytometry strategy parts the test and reference datasets, the AI device could naturally name the reference dataset as the “learning” dataset and use it to get familiar with the qualities of an untreated bacterium. Through ongoing examination with anti-toxin-treated cells, the device can recognize whether the microbes are helpless to the medication and could distinguish what extent of bacterial cells are safe in a blended opposition populace.
“Despite the fact that there was a misidentification blunder of under 10% in our work, there was a reasonable separation among helpless and safe cells in the span of 2 hours of anti-toxin treatment,” makes sense for Yoichiroh Hosokawa, one of the more senior creators in the gathering.
This work isn’t restricted to fast assessments of diseases in clinical settings. For example, drug discovery scientists could use it to quickly begin testing medication viability against any cell, as long as the cell reaction results in a change in dielectric properties. Impedance cytometry could become a staple of clinical and research labs before long.
More information: Tao Tang et al, Parallel Impedance Cytometry for Real-Time Screening of Bacterial Single Cells from Nano- to Microscale, ACS Sensors (2022). DOI: 10.1021/acssensors.2c01351
Tao Tang et al, Machine learning-based impedance system for real-time recognition of antibiotic-susceptible bacteria with parallel cytometry, Sensors and Actuators B: Chemical (2022). DOI: 10.1016/j.snb.2022.132698
Journal information: ACS Sensors