Joining AlphaFold2 with trial and computational strategies has assisted researchers in sorting out the human atomic pore complex’s engineering more meticulously than at any time in recent memory.
The human atomic pore complex (NPC) is a genuine sub-atomic monster, which sits on the film, isolating the core from the cytoplasm. It’s donut molded and fills in as both a passage and a designated spot for particles that move between the cytoplasm and the core. In this way, the NPC works with essential cycles in the cell, like quality articulation and interpretation. The atomic vehicle framework is also involved in a number of diseases, including neurodegenerative disorders, malignant growth, and viral contamination.
What’s the NPC’s construction? How are its proteins stuck together? How can it connect to the atomic layer? These and different inquiries have now been responded to by the Kosinski Group at EMBL Hamburg and the Center for Structural Systems Biology (CSSB), the Beck and Hummer Labs at the Max Planck Institute of Biophysics, and partners. They made the most incredibly complete model of the human NPC to date by consolidating the protein structure forecast program AlphaFold2 with procedures, for example, cryo-electron tomography, single molecule cryo-EM, and integrative display.
The human atomic pore complex (NPC) is a donut-shaped sub-atomic complex comprising of 30 unique proteins organized into ca. 1000 duplicates. It weighs 120 MDa, which on the cell scale is huge.
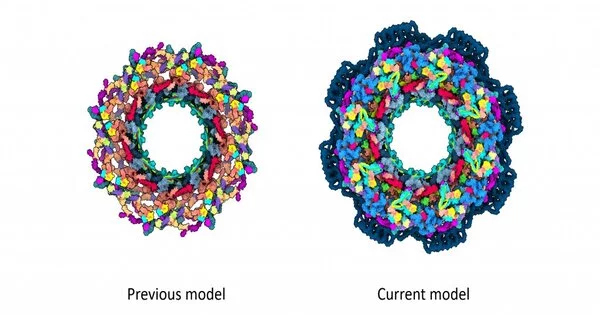
For primary researchers, the human NPC is a difficult yet thrilling 3D riddle, with around 30 unique proteins each present in different duplicates. This adds up to around 1000 unique pieces, which structure a round center with encompassing adaptable parts. Up to this point, the most reliable models of the human NPC center just covered 46% of the construction. Yet, presently, expanding on twenty years of past exploration in the field, researchers have made another model of the NPC structure that covers over 90% of its center.
While recently proposed NPC models had holes and contained a few proteins just in pieces, the new model eliminates quite a bit of this equivocalness.
“It’s similar to taking apart and reassembling an electronic equipment. There will always be a few screws missing, and you won’t know where they belong . We eventually got the majority of them in, and we now know exactly where they are, what they do, and how they do it.”
EMBL Group Leader Jan Kosinski.
“It resembles when you dismantle and reassemble an electronic gadget.” “There will be in every case a few screws left, and you simply don’t have the foggiest idea where they should be,” said EMBL Group Leader Jan Kosinski, who co-led the examination. “We have at long last figured out how to fit a large portion of them, and presently, we know precisely where they are, what they do, and how.”
Trial and error and man-made consciousness cooperate.
How did the researchers accomplish this? The key was to combine a few exploratory and computational strategies. This empowered the researchers to envision the NPC at various scales and levels of detail.
For instance, to demonstrate the general outline of the NPC, the scientists utilized cryo-electron tomography. With this strategy, they had the option to notice the NPC in its cell climate as opposed to in disconnection. More nuances of the unique protein building blocks were discovered by AlphaFold2, a DeepMind-created reasoning-based program that predicts protein structures.
“AlphaFold2 was a leading edge second for us,” said Agnieszka Obarska-Kosiska, the postdoc who played out the sub-atomic display. “Previously, we didn’t have the foggiest idea about the design of numerous proteins inside the NPC. You can’t solve a riddle when you don’t have the foggiest idea what the pieces resemble. In any case, AlphaFold2 joined with different methodologies empowered us to foresee those shapes.
To refine the image much further, the specialists utilized ColabFold, a variant of AlphaFold2 changed by established researchers to display cooperation between proteins. This permitted them to picture how the different unique pieces consolidate to frame more modest subcomplexes and how those subcomplexes are then stuck together to shape the NPC.
At last, they set up every one of the pieces utilizing the product Assembline, recently created by the Kosinski Group, and approved it for exploratory information.
The subsequent model was so finished and certain that it empowered the scientists to make time-settled sub-atomic reenactments that make sense of how the NPC proteins and the atomic layer cooperate to make a steady pore and how it answers mechanical signals.
Future headings
This work was a major leap forward for NPC research, yet there is still a ton left to investigate.
“This work epitomizes how, later on, primary science will embrace cell science to make nuclear models of ever bigger congregations of particles that carry out various roles in various pieces of the cell,” said Martin Beck. Gerhard Hummer concurs, “We can now imagine building a total powerful model of the NPC and reproducing the atomic vehicle in nuclear detail.”
The Kosinski Group will focus their future work on creating programmed strategies for incorporating underlying and microscopy information utilizing AlphaFold2 and their own product, Assembline. They intend to apply these ways to deal with concentrating on atomic cycles driving viral contamination.
The exploration was published in Science.