Cells are continually pursuing choices that lead to separation. For example, cells in an undeveloped organism pursue a progression of choices that decide if they will become neurons at times and muscle cells in others. How do cells settle on these choices?
Specialists at Texas A&M College and North Carolina State College are deciding the way in which cells work with dynamic cycles. Through this work, they desire to unequivocally gauge the centralization of explicit indispensable flagging proteins inside cell tissues. Furthermore, they will utilize the estimations to foster numerical models that can foresee and control cell separation.
This study was recently distributed in ACS Omega.
“We need to figure out separation choices, so we can at last tackle them,” said Dr. Gregory Reeves, an academic administrator in the Artie McFerrin Division of Compound Design at Texas A&M. “We are designing instruments to figure out cell separation and depict the cycles through conditions. To achieve these undertakings, we really want to figure out the centralization of the proteins in live tissues.
In any case, deciding the centralization of key flagging proteins can be very troublesome. To battle this issue, Reeves teamed up with North Carolina State College specialists who utilized a trial and insightful structure to foster blend and read exams. Blend and-read examines imply that basic reagents are set in mix with a lysed cell, considering glow recognition assuming that the objective protein is available.
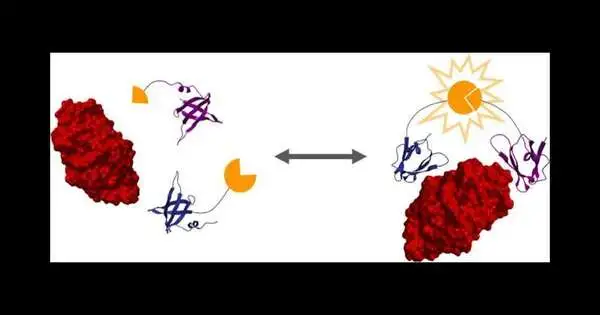
“When the target protein is bound by the two designed protein binders, it brings the two halves of luciferase together, resulting in bioluminescence, which we can utilize to collect measurements,”
Dr. Gregory Reeves, an associate professor in the Artie McFerrin Department of Chemical Engineering at Texas A&M.
The researchers then used a protein design method to create two proteins that tightly bind to an objective protein—in this case, lysozyme.These two protein folios are melded to two parts of luciferase, a compound that makes bioluminescence, as you would find in a firefly.
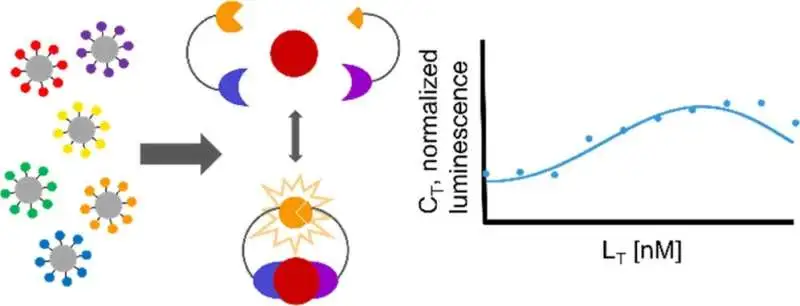
Graphical conceptual.
“At the point when the objective protein is limited by the two designed protein folios, it brings the two parts of luciferase together to make bioluminescence, which we can use to take estimations,” Reeves said.
Scientists from Reeves’ lab examined a numerical model of this strategy to foresee how much bioluminescence results from the limiting occasions, permitting them to decide the responsiveness of the measure. This, thusly, will assist specialists in acquiring a more profound understanding of how and why cells pursue separation choices.
The more extensive effects of this study incorporate utilizing this procedure to identify the presence of target proteins, like antibodies or upregulated disease markers, in a cell lysate.
“Different applications, which we will use in my lab, incorporate permitting us to neatly gauge a few proteins that couldn’t be estimated already in live tissues,” said Reeves.
The specialists likewise desire to additionally apply these strategies to different classes of atoms that are challenging to distinguish in live tissues, like mRNA.
This work is a joint effort with the lead creator, Nikki McArthur, and is supported by Dr. Balaji Rao, Dr. Carlos Cruz-Teran, and Apoorva Thatavarty from the Branch of Synthetic and Biomolecular Design at North Carolina State College.
More information: Nikki McArthur et al, Experimental and Analytical Framework for “Mix-and-Read” Assays Based on Split Luciferase, ACS Omega (2022). DOI: 10.1021/acsomega.2c02319
Journal information: ACS Omega