Watery bead development by fluid stage partition (or coacervation) in macromolecules is a hotly debated issue in life sciences research. Of these different macromolecules that structure beads, DNA is very fascinating in light of the fact that it is unsurprising and programmable, which are characteristics helpful in nanotechnology. As of late, the programmability of DNA has been utilized to build and manage DNA drops framed by coacervation of succession planned DNA.
A group of researchers at Tokyo University of Technology (Tokyo Tech) led by Prof. Masahiro Takinoue has fostered a computational DNA bead with the capacity to perceive explicit mixes of synthetically combined microRNAs (miRNAs) that go about as biomarkers of growth. Involving these miRNAs as sub-atomic information, the drops can give a DNA rationale figure yield through actual DNA bead stage partition. Prof. Takinoue makes sense of the requirement for such examinations: “The uses of DNA beads have been accounted for in cell-roused microcompartments.” Despite the fact that organic frameworks manage their capacities by consolidating biosensing with atomic coherent calculation, no writing is accessible on the combination of DNA drop with sub-atomic registering. ” Their discoveries were published in Advanced Functional Materials.
“We can employ a DNA droplet that can combine and process many inputs and outputs in early illness diagnosis and medicine delivery systems if one can be produced. Our current research also serves as a springboard for future work on artificial intelligence and molecular rob otics.”
Prof. Takinoue
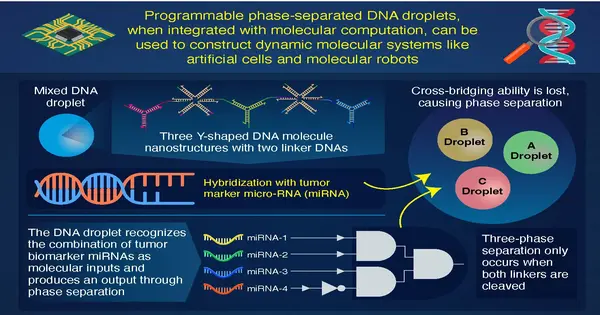
Fostering this DNA bead required a progression of trials. In the first place, they planned three sorts of Y-molded DNA nanostructures considered Y-themes A, B, and C with three tacky finishes to make A, B, and C DNA drops. Ordinarily, comparable drops gather as one naturally, while to join divergent drops, a unique “linker” particle is required. In this way, they utilized linker particles to get the A drop together with the B and C drops; these linker particles were called AB and AC linkers, separately.
In their most memorable trial, they assessed the “AND” activity in the AB bead blend by presenting 2 info DNAs. In this activity, the presence of information is recorded as 1, while its nonattendance is recorded as 0. The stage partition of the AB drop combination happened exclusively at (1,1), meaning when both information DNAs are available, recommending effective utilization of the AND activity. Following this review, the researchers chose to present bosom disease growth markers, miRNA-1 and miRNA-2, to the AC bead blend as contributions to the AND activity. The AND activity was effective, inferring that the computational DNA bead distinguished the miRNAs.
In the resulting tests, the group showed synchronously AND as well as NOT activities in the AB blend with miRNA-3 and miRNA-4 bosom malignant growth biomarkers. Ultimately, they made an ABC drop blend and presented all the 4 bosom disease biomarkers to this arrangement. The stage partition in ABC bead relied upon the linker cleavage, bringing about a two-stage detachment or a three-stage division.
This property of the ABC drop empowered the scientists to show the capacity to identify a bunch of known malignant growth biomarkers or distinguish markers of 3 illnesses all at once. Prof. Takinoue, who is also the related creator, believes that computational DNA drops have enormous potential. As per him, “On the off chance that a DNA drop can be created which can coordinate and handle numerous data sources and results, we can involve it in early sickness identification as well as medication conveyance frameworks.” Our momentum concentrate likewise goes about as a steppingstone for research in creating wise fake cells and sub-atomic robots. “