The layer that encases an organic cell isn’t just a hindrance; it is packed with proteins associated with a wide range of basic natural capabilities. To truly comprehend what layer proteins are doing and how, analysts need to realize how they’re coordinated and the way that they communicate with each other. However, it is trying to uncover that data.
Yale scientists have now fostered another microscopy technique called Local nanoBleach that defeats the primary difficulties to figuring out film protein association, remembering the trouble of concentrating on these layers without upsetting the local climate and cutoff points to the goal of light magnifying instruments normally used to concentrate on them.
What’s more, to show the viability of the new strategy, they effectively applied it to a natural problem—relating to proteins engaged with the improvement of pancreatic diseases and how they may be focused on for treatment—that has stayed strange for quite a long time.
“We get the best of both worlds with our method: we keep the native membrane environment while having very high spatial and single-molecule resolution. When we used our approach, we discovered that KRas occurs in similar proportions as dimers and monomers. Dimers rise and monomers decrease when KRas is altered, as in pancreatic cancer.”
Moitrayee Bhattacharyya, assistant professor of pharmacology at Yale School of Medicine.
They depict the new technique and its benefits in another review distributed in Nature Nanotechnology.
Strategies ordinarily utilized in the investigation of film protein association require eliminating the local layer climate encompassing proteins and afterward placing segregated proteins of premium into conditions that copy but don’t completely recreate the intricacy of the genuine cell film, said Moitrayee Bhattacharyya, right-hand teacher of pharmacology at Yale Institute of Medicine and senior creator of the review. This methodology, Bhattacharyya said, eliminates significant setting since the proteins associate with the particles that encompass them.
Second, light magnifying instruments, which are regularly used to notice protein association, don’t have the goal expected to decide if proteins close to one another are really communicating or are just neighbors on layers.
Last, how much of a specific protein found in a cell layer might be excessively low or excessively high for the current techniques for study? In those cases, specialists need to make changes; they either need to imitate proteins that, in their normal state, are too few or separate out proteins from tests where there are too many. Yet, this again can eliminate significant information about the normal condition of proteins as they sit and work in cell layers.
“In a perfect world, we would have a technique that would work with any endogenous degree of cell film protein articulation,” said Bhattacharyya.
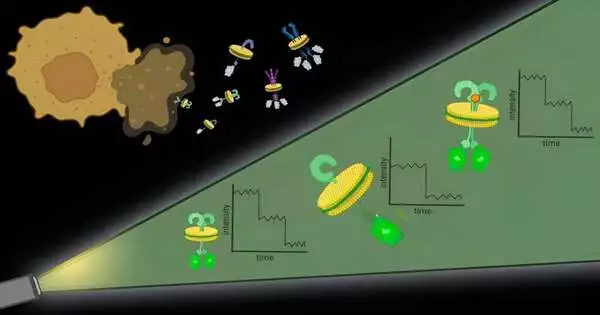
To handle the primary test, the analysts utilized specific atoms and sorts of polymers to basically finish off protein edifices with their encompassing cell film. “It resembles the cell film, which is a sheet of treat batter, and the polymers are dough shapers,” said Bhattacharyya.
These pieces of protein with an encompassing cell layer, named local nanodiscs, are roughly 10 nanometers in measurement, little enough that any proteins contained in the nanodisc are logically communicating, which tends to the subsequent test. Further, this approach works with any protein in the cell layer, permitting scientists to notice proteins at their normal levels in local films.
Once the nanodiscs are created, scientists can utilize quite a few normally utilized methods to focus on a specific protein of interest. They then, at that point, measure the proteins in each nanodisc with the assistance of fluorescent particles connected to them.
A methodology offers high spatial goals without the requirement for specific equipment, said Bhattacharyya.
“This work presents another method to grasp how layer proteins—which address around 60% of medication targets—gather into useful units on or inside the local lipid bilayer,” said Gerard Walker, co-first creator of the paper and an alumni understudy in Bhattacharyya’s lab.
To show how this strategy may be applied, the specialists required a decades-long banter in science. A protein called KRas is changed in over 90% of human pancreatic tumors, generating huge clinical and helpful interest. Whether KRas subunits meet up to frame dimers (two units) or oligomers (multiple units) on cell layers has remained the focal point of longstanding examination.
Studies, be that as it may, have created clashing discoveries. Creature and cell studies, which need point-by-point atomic goals, show proof that KRas units meet up on cell layers. In the mean time, biophysical examinations, which don’t hold the local layer around proteins, have found KRas stays in single units, or monomers.
“With our strategy, we outwit the two universes,” said Bhattacharyya. “We hold the local film climate, and we have extremely high spatial and single-particle goals. At the point when we applied our technique, we found that KRas existed as dimers and monomers in comparable amounts. However, when KRas is changed, as in pancreatic diseases, dimers increment and monomers decline.”
The tracking down features the significance of the local cell layer for figuring out film proteins and recognizes an objective—decreasing KRas dimerization—for disease treatment. This is only one of numerous ways this technique could be utilized to comprehend the job of layer protein association in illness, Bhattacharyya said.
“It’s truly remunerating to see Local nanoBleach previously being effectively applied to a wide assortment of squeezing organic inquiries in the Bhattacharyya lab and then some,” said Caroline Brown, co-first creator of the review and a Ph.D. up-and-comer in the lab of co-creator Kallol Gupta, aide teacher of cell science.
Layer proteins make up 33% of each of the proteins in the human body, and this approach can be utilized to concentrate on any of them, said Bhattacharyya.
“It’s an overall procedure,” she said. “There’s actually no restriction.”
Going ahead, Bhattacharyya and her partners desire to stretch out this way to deal with concentrating on protein association in the layers of different organelles and structures, such as mitochondria, that are held inside cells.
More information: Gerard Walker et al, Oligomeric organization of membrane proteins from native membranes at nanoscale spatial and single-molecule resolution, Nature Nanotechnology (2023). DOI: 10.1038/s41565-023-01547-4