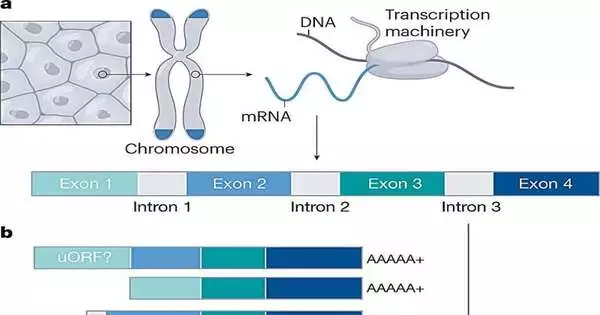
The human genome, from one end to another, has been sequenced, meaning researchers have overall distinguished the greater part of the almost 20,000 protein-coding qualities. Be that as it may, a global gathering of researchers noticed there’s more work to be finished. The researchers call attention to the fact that, despite the fact that we have almost combined the characters of the 20,000 qualities, the qualities can be sliced and joined to make roughly 100,000 proteins, and quality specialists are a long way from settling on what those 100,000 proteins are.
The gathering, which met the previous fall at the Cold Spring Harbor Research facility in New York, has now distributed an aid for focusing on the following stages in the work to finish the human quality “list.”
“Numerous researchers have been dealing with endeavors to completely figure out the human genome, and it’s significantly more troublesome and complex than we suspected,” says Steven Salzberg, Ph.D., Bloomberg Recognized Teacher of Biomedical Designing, Software Engineering, and Biostatistics at The Johns Hopkins College. “We have given a condition of the human quality index and an aid on what’s expected to finish it.”
“Many scientists have been working on attempts to fully comprehend the human genome, and it’s far more difficult and complex than we expected. We’ve published an update on the condition of the human gene catalog, as well as a guidance on what’s needed to finish it.”
Steven Salzberg, Ph.D., Bloomberg Distinguished Professor of Biomedical Engineering, Computer Science, and Biostatistics at The Johns Hopkins University.
Salzberg, alongside Johns Hopkins biomedical designer and academic partner Mihaela Pertea, Ph.D., M.S., M.S.E., postdoctoral analyst Lagers Varabyou, and 19 different researchers, offered points of view on the human quality inventory on October 4 in the journal Nature.
The researchers express that while the last rundown of protein coding qualities is almost finished, they have not yet completely classified the range of ways that a quality can be cut, or grafted, coming about in “isoforms” of proteins that are somewhat unique. Some protein isoforms won’t influence the protein’s capability; however, some might be sufficiently different to bring about an expanded risk for a specific quality, condition, or sickness.
To finish the index, the researchers propose an extensive gander at how every quality is communicated into useful and nonfunctional proteins and the three-layered state of those proteins.
The researchers likewise propose an emphasis on classifying non-coding RNA qualities. RNA is the hereditary material that is translated by DNA and follows a sub-atomic way to make proteins. Rather than proteins, non-coding RNA qualities encode different kinds of sub-atomic material that carries out a cell role.
At long last, the worldwide gathering accentuates the significance of upgrading regularly utilized information bases of quality varieties that cause sickness and infection, further developing clinical lab principles for commenting on DNA sequencing results, and growing new innovation to empower more powerful and exact strategies to coordinate the wide cluster of proteins with their quality items.
More information: Paulo Amaral et al, The status of the human gene Catalog, Nature (2023). DOI: 10.1038/s41586-023-06490-x