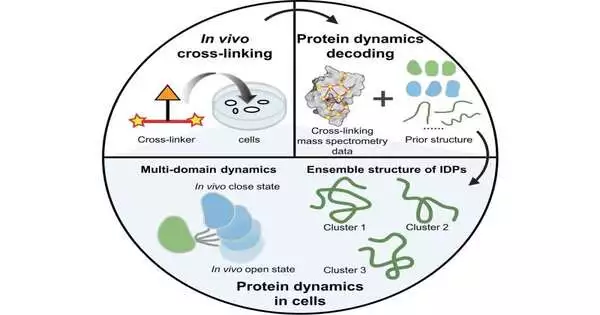
Protein elements assume a significant role in different capabilities. The intracellular climate essentially impacts protein elements, especially inherently scattered proteins (IDPs).
An examination bunch drove Prof. Zhang Lihua from the Dalian Establishment of Compound Physical Science (DICP) of the Chinese Foundation of Sciences (CAS), as a team with Assoc. Prof. Gong Zhou from the Accuracy Estimation Science and Innovation Development Exploration Foundation of CAS, has proposed a technique involving in-vivo substance cross-connecting and mass spectrometry (in-vivo XL-MS) to disentangle the unique design of proteins inside cells.
In-vivo XL-MS has potential for examining the powerful construction of proteins inside cells because of its high throughput, high responsiveness, and low prerequisites for protein immaculateness.
This study was distributed in Angewandte Chemie Global Version on July 5.
AlphaFold, a formerly proposed protein structure forecast strategy, consolidates novel brain network models and prepares methods that depend on the development of primary examples and the coordination of physical and mathematical restrictions.
In this review, the scientists used the earlier construction from AlphaFold2 as earlier data and consolidated in-vivo XL-MS information with different underlying estimation techniques to survey the similarity among designs and cross-connecting data. They remade the powerful designs of different proteins inside cells.
They zeroed in on multi-space proteins and proposed a technique to regard the areas in general, using XL-MS information between the areas to display the powerful designs of proteins inside cells. They described the unique designs of three multi-space proteins, in particular calmodulin, hnRNP A1, and hnRNP D0.
Concerning IDPs, they presented two correlative primary portrayal procedures: one included straightforwardly changing over XL-MS information into distance imperatives for IDP structure estimation, while the other technique utilized impartial testing utilizing all-iota sub-atomic element reenactments, followed by assessment and determination of the inspected structures in light of XL-MS information. By utilizing these two procedures, they decoded the outfit conformities of two exceptionally versatile, high-portability bunch proteins inside cells, HMG-I/Y and HMG-17.
“Our review offers specialized help for a more profound comprehension of the sub-atomic components fundamental protein usefulness in the cell microenvironment,” said Prof. Zhang.
More information: Beirong Zhang et al, Decoding Protein Dynamics in Cells Using Chemical Cross‐Linking and Hierarchical Analysis, Angewandte Chemie International Edition (2023). DOI: 10.1002/anie.202301345. On BioarXiv: DOI: 10.1101/2023.03.21.533582