A College of Rhode Island teacher of Sea Designing and Oceanography, alongside a multidisciplinary research group from various organizations, effectively showed new innovations that can get protected tissue and high-goal 3D pictures promptly after experiencing probably the most delicate creatures in the profound sea.
URI Teacher Brennan Phillips, the important specialist on the undertaking, and a group of 15 scientists from six establishments, including URI, have shown that it is feasible to shave a very long time from the most common way of deciding if a new or interesting animal variety has been found. The consequences of their work are distributed today (Jan. 17) in the journal Science Advances.
Roboticists, sea designers, bioengineers, and marine and sub-atomic scientists from URI’s Branch of Sea Designing; the Bigelow Lab for Sea Sciences in East Boothbay, Maine; the School of Designing and Applied Sciences at Harvard College; Monterey Narrows Aquarium Exploration Organization (MBARI) in California; Dad Counseling, an overall firm that spotlights on development; and the Division of Innate Sciences at Baruch School, City College of New York, made up the group. The paper addresses five years of examination.
“Currently, researchers confront a difficult task when attempting to describe what they think to be a new species. The current method involves capturing a specimen, which is extremely challenging because many of these organisms are so delicate and tissue-thin that they may not be able to be collected at all. If you successfully collect an animal, you can then preserve it in a jar.”
Professor Brennan Phillips, the principal investigator on the project.
Progressive headways in submerged imaging, advanced mechanics, and genomic sequencing have reshaped marine investigation, the review affirms. The examination shows that, promptly after an experience with a remote ocean creature, it is feasible to catch definite estimations and movements of the creature, get a whole genome, and produce an exhaustive rundown of qualities being communicated that highlight their physiological status in the profound sea. The consequence of this rich computerized information is a “cybertype” of a solitary creature, as opposed to a physical “holotype” that is customarily tracked down in historical center assortments.
A revolving incited dodecahedron (RAD-2) exemplifies a holoplanktonic polychaete (Tomopteris, a marine worm). Video from a remotely operated vehicle (ROV), a Subastian science camera, and the Schmidt Sea Establishment. Credit: Schmidt Sea Organization
“As of now, to portray what they accept as another species, they face a burdensome cycle,” Phillips said. “How it is done now is that you catch an example, which is undeniably challenging on the grounds that a ton of these creatures are so fragile and tissue-slim, and it’s logical you will be unable to gather them by any means. Be that as it may, on the off chance that you effectively gather a creature, you protect it in a container.
“Then starts a long course of genuinely carrying that example to various assortments all over the planet where it is contrasted with existing living beings. After quite a while, in some cases as long as 21 years, researchers might arrive at the conclusion that this is another species.
“Once more, these are remote oceans, dainty little wisps of creatures. The ongoing work process isn’t fitting. It’s a significant motivation behind why we have so many undescribed species in the sea.”
Data acquired from the review—and others that follow—could be valuable for termination counteraction studies, as it gives an abundance of data from a solitary example acquired during a solitary experience. The work additionally answers the developing call among specialists for humane assortment, which limits damage to creatures by utilizing trend-setting innovations to gather data. Future examinations and improvements could consider total sweeps and inventories of life in the remote ocean inside a catch-and-delivery system.
“The vision was: The means by which could a sea life scholar work to all the more likely comprehend and interface with remote ocean life many years or hundreds of years into what was in store?” David Gruber is a recognized teacher of science at Baruch School, City College of New York, and a Pilgrim with Public Geographic Culture. “This is a show of how an interdisciplinary group could function cooperatively to give a gigantic measure of new data on remote ocean life after one brief experience.
“A definitive objective is to go on down this way and refine the innovation to be basically as negligibly obtrusive as could really be expected—similar to a specialist’s examination in the remote ocean. This approach is turning out to be progressively significant, with current termination rates being multiple times higher than foundation annihilation rates.”
Phillips said, on the grounds that gathering these examples has forever been hard, that there are some remote ocean species that still can’t seem to be distinguished. “At the point when you see environmental change and remote ocean mining and their expected impacts, it is disrupting,” Phillips said. “You understand you don’t have a full benchmark of animal varieties, and you may not understand what you’ve lost before it’s past the point of no return. If you have any desire to understand what has been there before it’s gone, this is a better approach to doing that.”

The ROV SuBastian with a turning-impelled dodecahedron (RAD-2) mounted on the front is going to be brought down into the ocean. Credit: Brennan Phillips
The mission was led by the exploration vessel Falkor and included two campaigns off the banks of Hawaii and San Diego in 2019 and 2021. The group gathered upwards of 14 safeguarded tissue tests a day, along with terabytes of quantitative computerized symbolism. Together, the review gave:
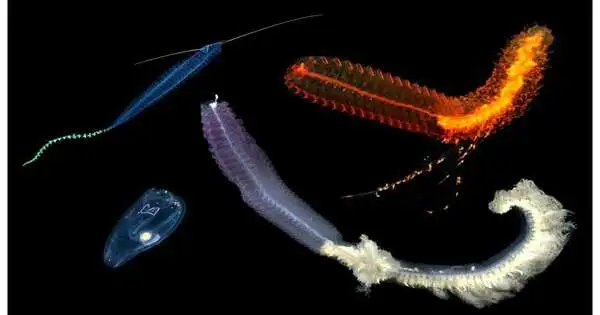
The first complete collected and commented on transcriptome (qualities being made in the creatures’ environment) of Pegea tunicate, a marine invertebrate creature;
Subtleties of the sub-atomic premise of natural detection of a holoplanktonic Tomopteris polychaete (marine worm), which goes through its whole time on earth in the water segment;
Subtleties of the full transcriptomes of two siphonophores (coagulated zooplankton made out of particular parts filling together in a chain): Erenna sp., Marrus claudanielis, as well as the Pegea tunicate and Tomopteris polychaete;
Full morphological (structure and design) portrayals utilize computerized imaging of every creature while at depth.
The lead creator of the paper, John Consumes, a senior exploration researcher at Bigelow Lab, directed the genomic investigation on four creatures tested at profundities of very nearly 4,000 feet.
“What we had the option to accomplish with these creatures is amazing,” Consumes said. “For my purposes, this is best found in the succession information we created for the Tomopteris worm: We caught it while it was investigating its current circumstance and had the option to construe that it was checking the water involving two long tangible bristles close to its head for’sweet’ tastes: reasonable sugars related to prey, and potentially for smelling salts: a side-effect of its run of the mill prey.
“With that data, we can imagine how it chases by following substance trails in its untamed water territory,” Consumes said. “I don’t believe that would have been conceivable without the creative innovation designed and utilized by the specialists in the group that permitted total protection of the data from the creatures not long after an experience.”
Consumes said one more review with Gruber saw what catch strategies mean for jellyfish ribonucleic corrosive, known as RNA, one of the structural blocks of life. That arrangement of data can begin to change after around 10 minutes of unpleasant circumstances, even with a delicate assortment. The Planning the Future innovations conquer this by safeguarding the data before the creature’s cells begin to answer pressure, as indicated by Consumes.
“We likewise found that three of the creatures we caught have colossal genomes, each having almost multiple times the DNA in a cell compared with us,” Consumes said. “For the fourth, with an all the more unobtrusively estimated genome (around 3% the size of a human genome), we had the option to utilize front-line sequencing techniques to fabricate the most strong and complete genome of a salp to date.”
Harvard and URI brought to the mission a rotating incited collapsing dodecahedron (RAD-2), an inventive origami-roused mechanical exemplification gadget that gathered creature tissue tests and immediately protected that tissue at profundity.
“We are seeing the effect of new kinds of marine robots for midwater and remote ocean investigation,” said roboticist Robert Wood, the Harry Lewis and Marlyn McGrath Teacher of Designing and Applied Sciences at Harvard College. “In addition to the fact that robots are going to spots that are troublesome or unimaginable for people to come to, our gadgets research, cooperate with, and gather examples utilizing a delicate touch… or no touch by any stretch of the imagination.”
Imaging frameworks from MBARI’s Bioinspiration Lab that incorporated a laser-filtering imaging gadget called DeepPIV and a three-layered lightfield camera called EyeRIS empowered the estimation and recreation of the three-layered morphology, or body shape, of the creatures right at home.
“We can’t safeguard what we don’t yet completely comprehend. High-level imaging advances can speed up our endeavors to report the variety of life in the sea. The quicker we can index marine life, the better we can evaluate and follow the effect of human activities like environmental change and mining on sea conditions,” said Kakani Katija, bioengineer and chief specialist of the Bioinspiration Lab at MBARI.
“We have these remotely operated vehicles out there with cutting-edge imaging frameworks, which can make a three-layered model after a couple of moments,” Phillips said. “We had the option to move toward a little jellyfish in no time, gather high-goal 3D pictures to the control room, and our group had the option to tell in practically no time that the limbs were precisely 5 millimeters in length. Then, we had very much saved tissue tests of similar creatures inside merely minutes.”
More information: John Burns et al, An in situ digital synthesis strategy for the discovery and description of ocean life, Science Advances (2024). DOI: 10.1126/sciadv.adj4960. www.science.org/doi/10.1126/sciadv.adj4960